
We have developed a new strategy termed "potency coherence analysis" that leverages the drug potency dimension in decryptM to decode the kinases that shape the human phosphoproteome.
Read more:

Koina aims to transform how #proteomics uses machine learning. You no longer need to be a tech wizard to use ML and now can easily run #ML models. Integrated with FragPipe, Skyline and EncyclopeDIA!
www.nature.com/articles/s41...

Koina aims to transform how #proteomics uses machine learning. You no longer need to be a tech wizard to use ML and now can easily run #ML models. Integrated with FragPipe, Skyline and EncyclopeDIA!
www.nature.com/articles/s41...
We're excited to spend the next days reuniting with old friends and making new connections. See you there!

We're excited to spend the next days reuniting with old friends and making new connections. See you there!
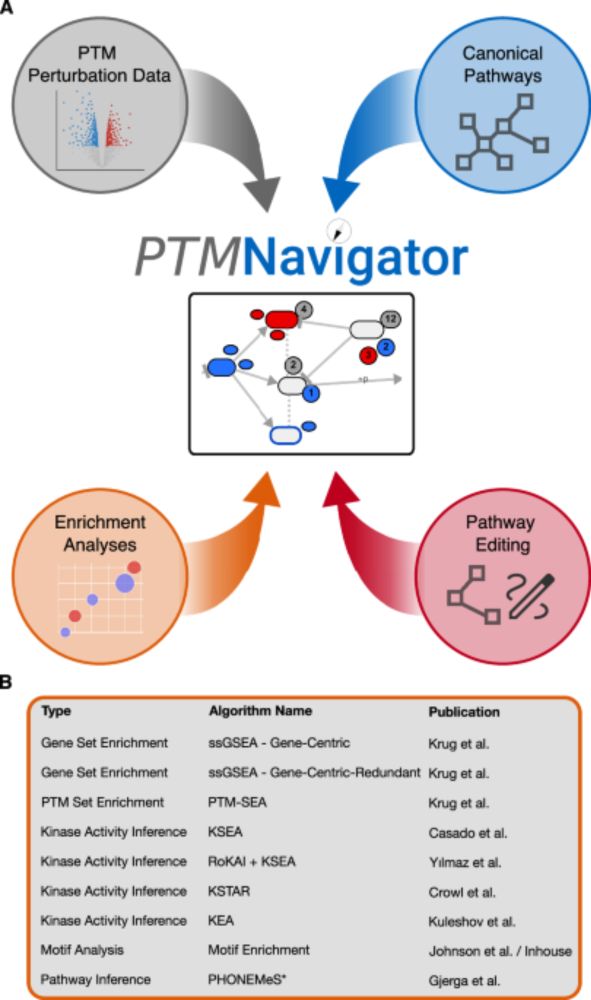
We cover databases, enrichment tools, software for pathway reconstruction, and full-fledged platforms that help to interpret high-throughput PTM datasets.
Check it out here: doi.org/10.1002/pmic...

We cover databases, enrichment tools, software for pathway reconstruction, and full-fledged platforms that help to interpret high-throughput PTM datasets.
Check it out here: doi.org/10.1002/pmic...


www.ls.tum.de/en/ls/public...
If you haven't had the chance to read the publication, you can check it out here: doi.org/10.1016/j.ce...

www.ls.tum.de/en/ls/public...
If you haven't had the chance to read the publication, you can check it out here: doi.org/10.1016/j.ce...
www.science.org/doi/10.1126/...
(1/4)
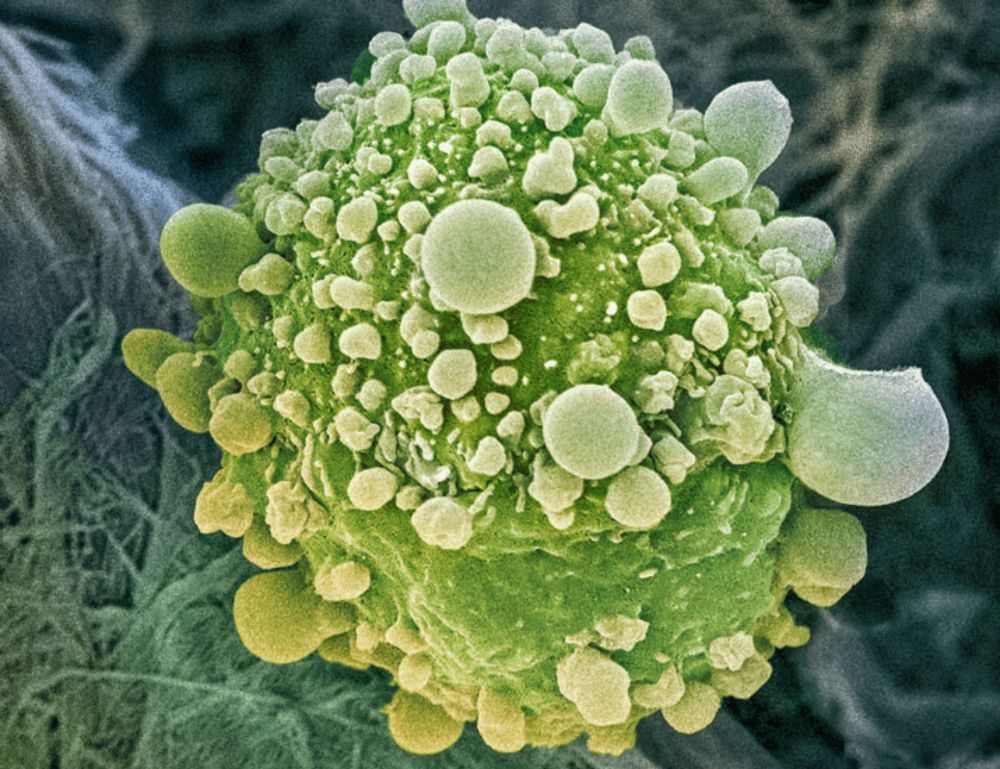
www.science.org/doi/10.1126/...
(1/4)
Apply now if you're interested in working with us!
Apply now if you're interested in working with us!
Go #TeamMassSpec!

Go #TeamMassSpec!
If you're attending today, make sure to check out @msleelab.bsky.social's poster on citrullination in multiple sclerosis. Sophia 'Lapo' Laposchan is excited to see you there!

If you're attending today, make sure to check out @msleelab.bsky.social's poster on citrullination in multiple sclerosis. Sophia 'Lapo' Laposchan is excited to see you there!
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...
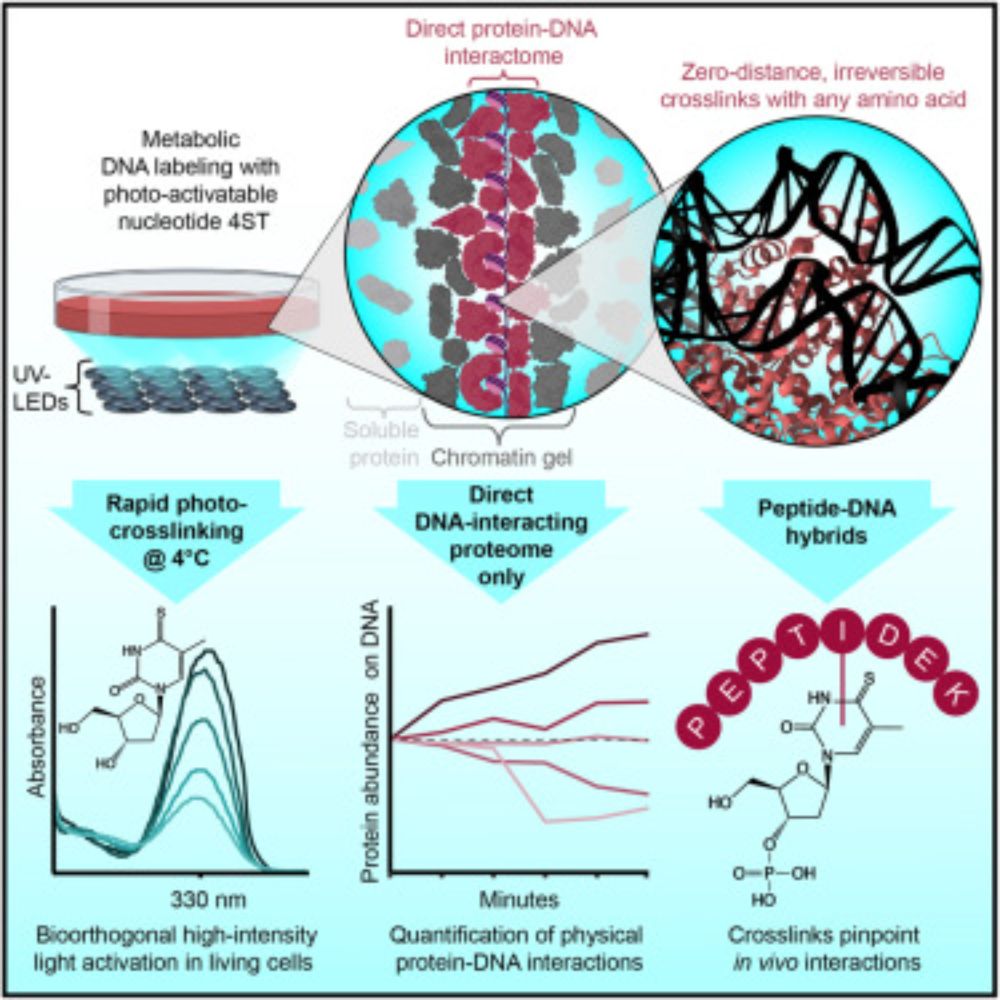
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b

Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b
With #CHIMERYS, we can now directly compare DDA and DIA data — 🍎 to 🍎 finally made possible.
doi.org/10.1038/s41592-025-02663-w
#KusterLab #WilhelmLab #MSAID #Proteomics
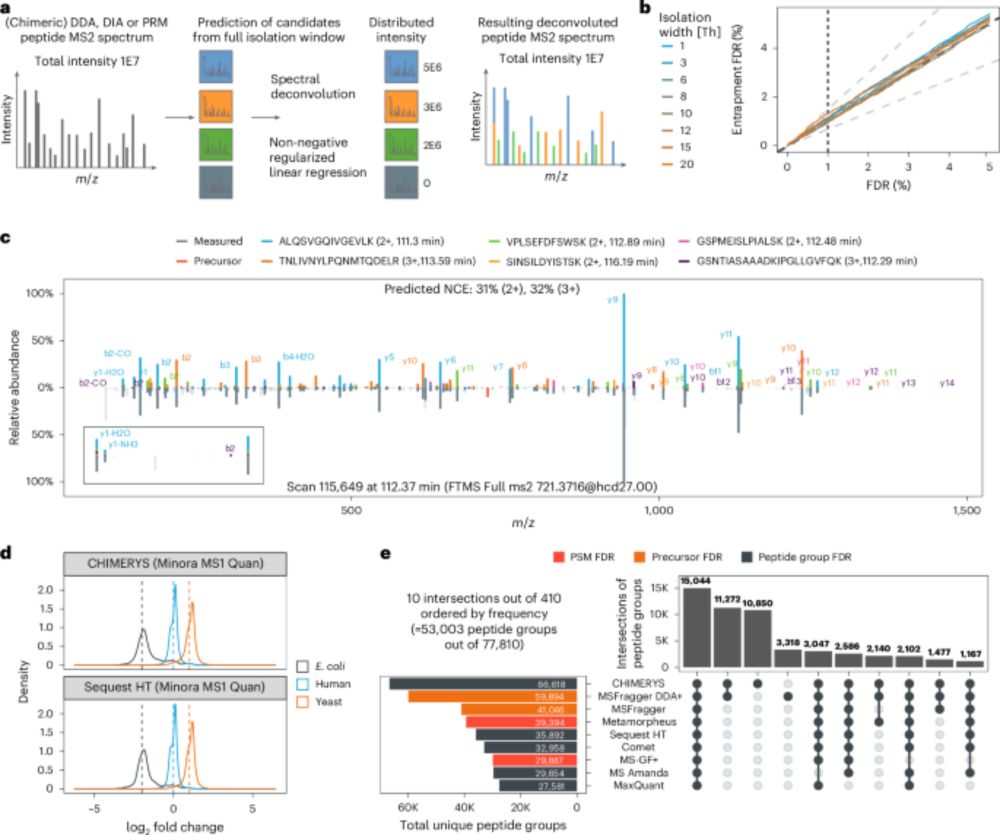
With #CHIMERYS, we can now directly compare DDA and DIA data — 🍎 to 🍎 finally made possible.
doi.org/10.1038/s41592-025-02663-w
#KusterLab #WilhelmLab #MSAID #Proteomics
metabolic mutations drive bacterial antibiotic resistance evolution, prior knowledge for GRN inference, HPA drugs fail mood disorder clinical trials
Cover: ATR kinase inhibitors work together to overcome chemoresistance @kusterlab.bsky.social

metabolic mutations drive bacterial antibiotic resistance evolution, prior knowledge for GRN inference, HPA drugs fail mood disorder clinical trials
Cover: ATR kinase inhibitors work together to overcome chemoresistance @kusterlab.bsky.social

📄 doi.org/10.1038/s414... (1/6)
📄 doi.org/10.1038/s414... (1/6)
🚨 New Research Alert! 🚨
Excited to share our latest work in @embojournal.bsky.social
"Towards routine proteome profiling of FFPE tissue: insights from a 1,220-case pan-cancer study" ✨
doi.org/10.1038/s443...
📌 Key Highlights:

🚨 New Research Alert! 🚨
Excited to share our latest work in @embojournal.bsky.social
"Towards routine proteome profiling of FFPE tissue: insights from a 1,220-case pan-cancer study" ✨
doi.org/10.1038/s443...
📌 Key Highlights:

