

James C Bowden, Sergey Levine @jlistgarten.bsky.social
arxiv.org/abs/2511.03032
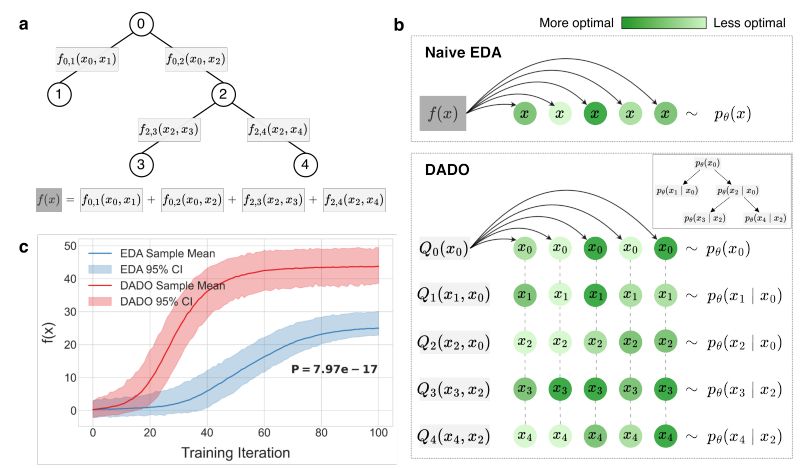

James C Bowden, Sergey Levine @jlistgarten.bsky.social
arxiv.org/abs/2511.03032
www.biorxiv.org/content/10.1...


www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...


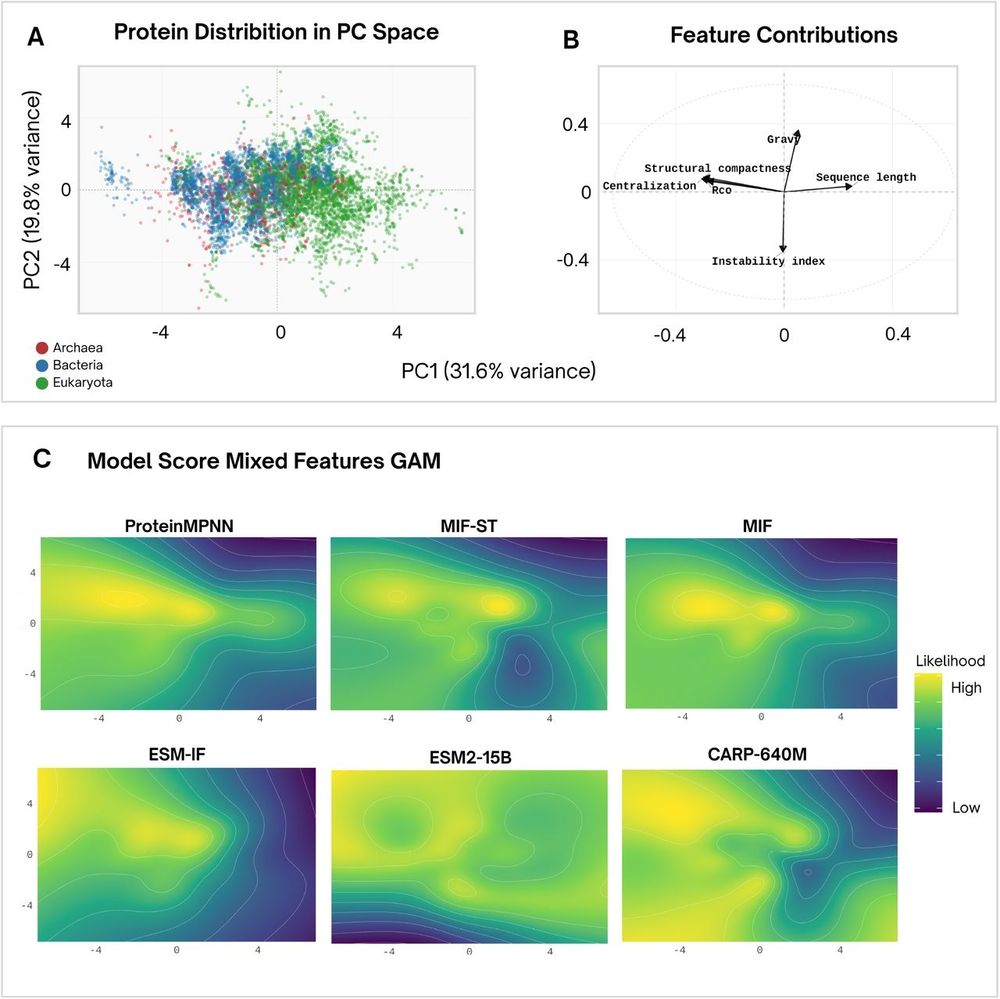

www.biorxiv.org/content/10.1...
@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...



@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...
Arjuna Subramanian, Matt Thomson
www.biorxiv.org/content/10.1...


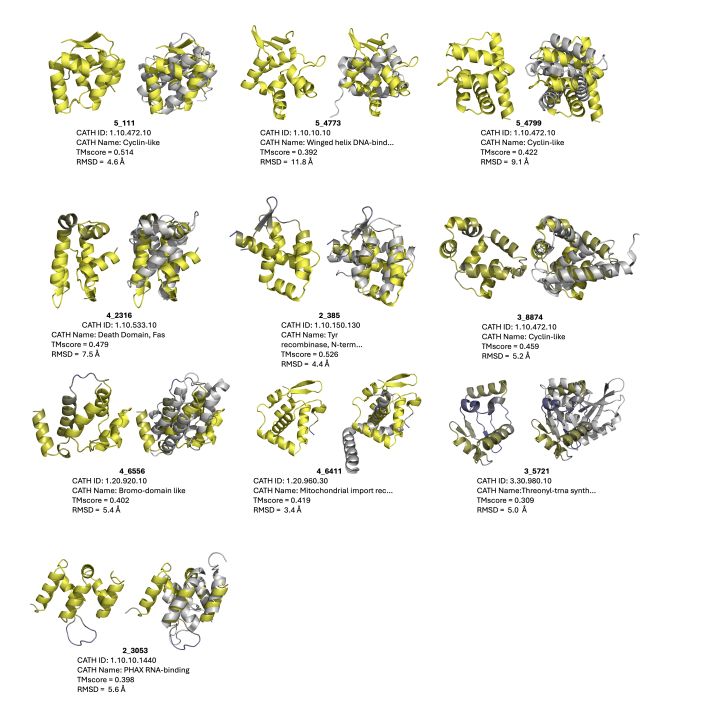
Arjuna Subramanian, Matt Thomson
www.biorxiv.org/content/10.1...
@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...
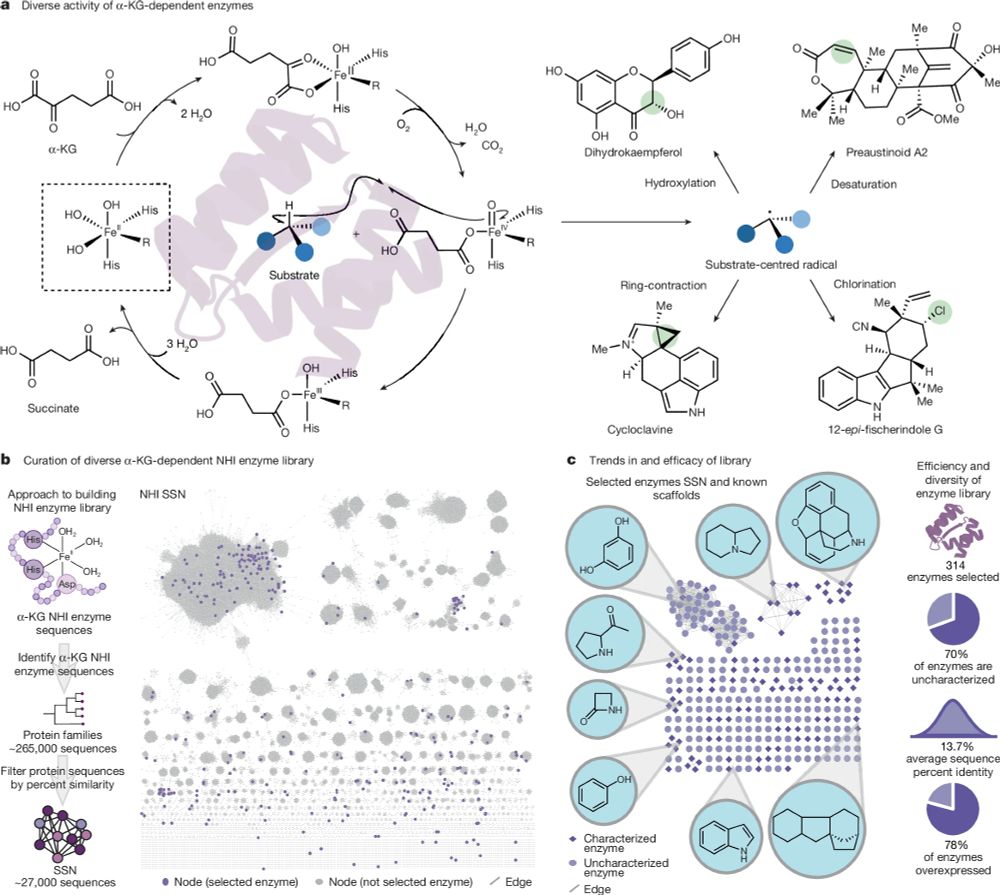

@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...
@synbiogaolab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...



@synbiogaolab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
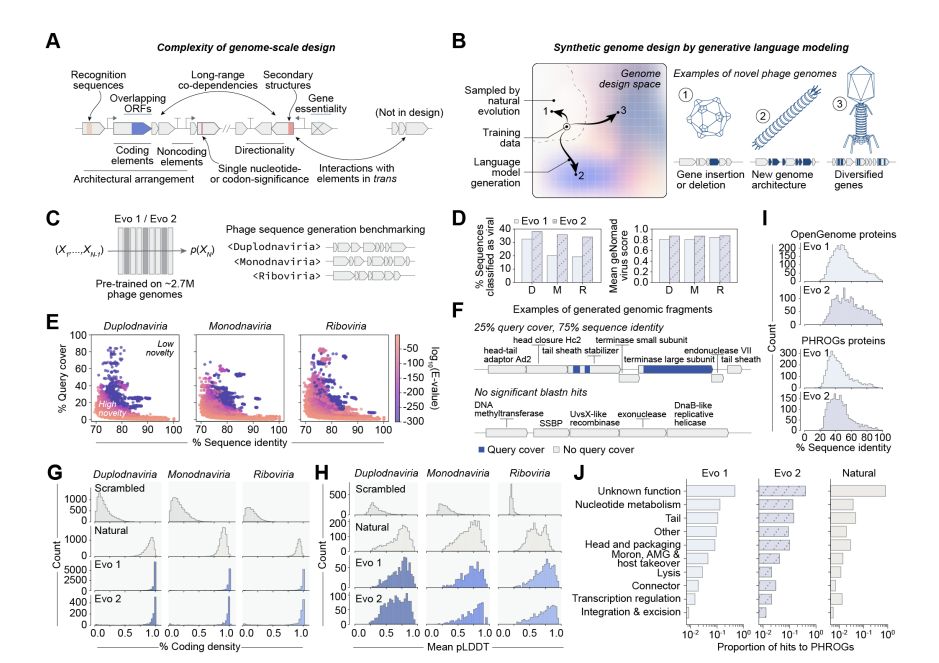

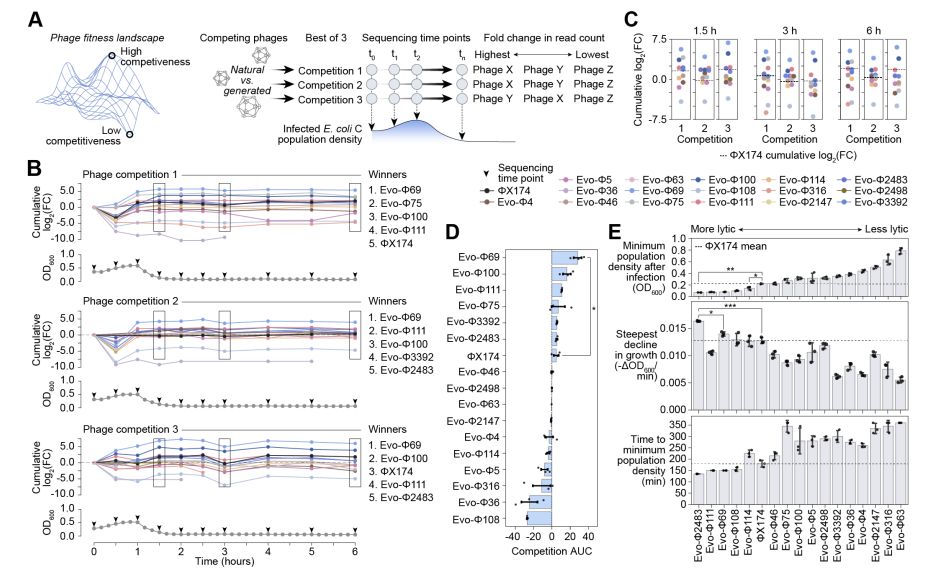

@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
@panhammarstrom.bsky.social @patrickbryant1.bsky.social
www.biorxiv.org/content/10.1...
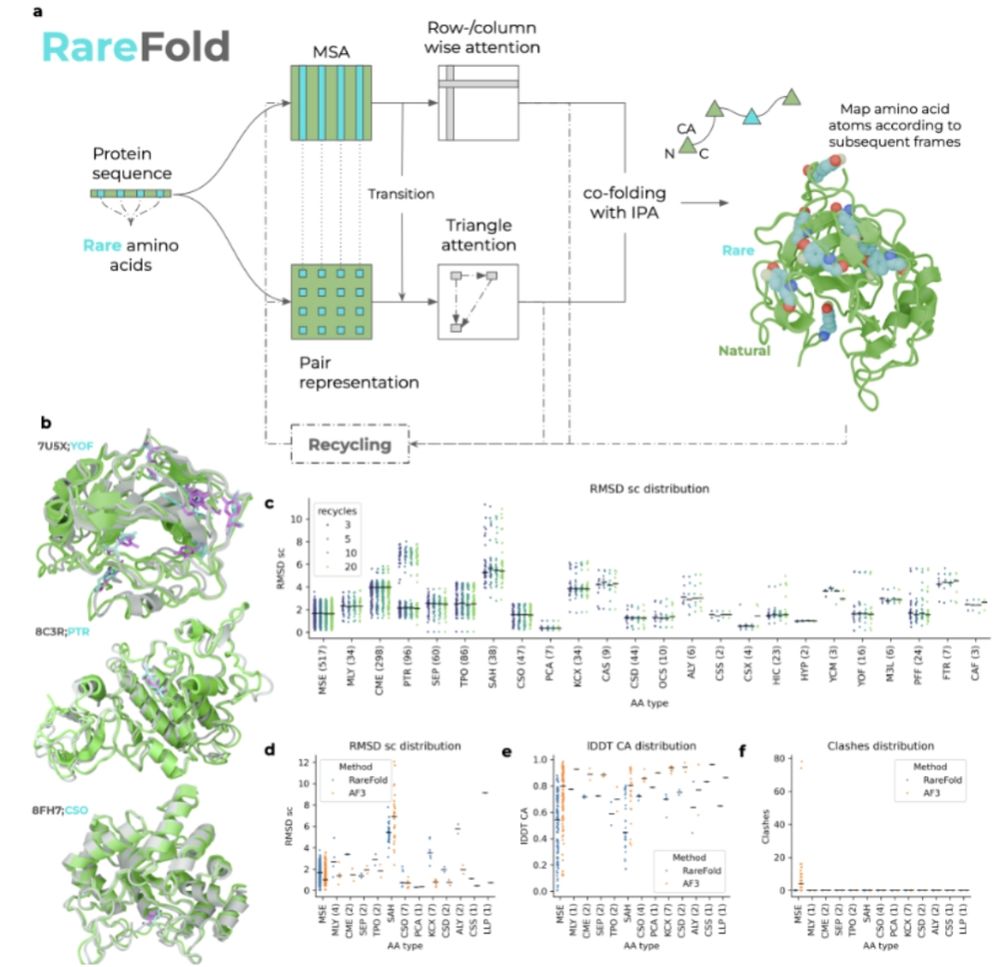

@panhammarstrom.bsky.social @patrickbryant1.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...


www.biorxiv.org/content/10.1...
@jsunn-y.bsky.social @francescazfl.bsky.social Yueming Long @francesarnold.bsky.social
www.cell.com/cell-systems...

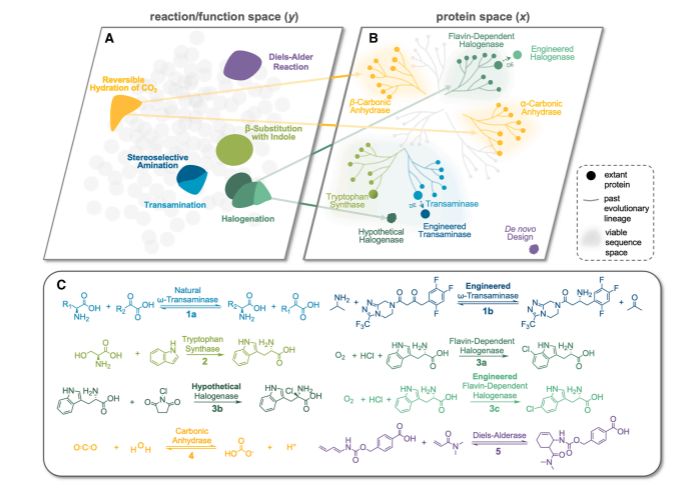
@jsunn-y.bsky.social @francescazfl.bsky.social Yueming Long @francesarnold.bsky.social
www.cell.com/cell-systems...

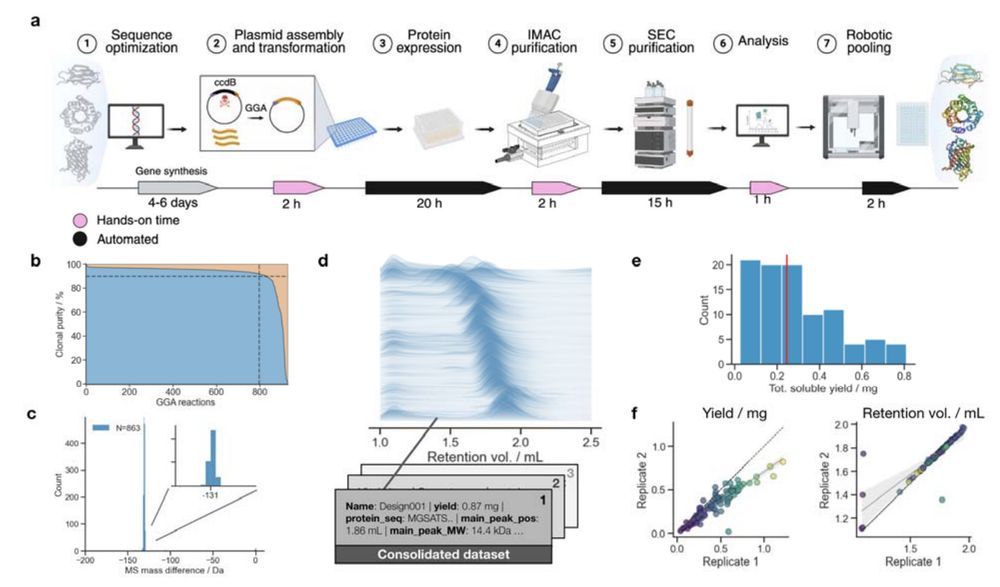
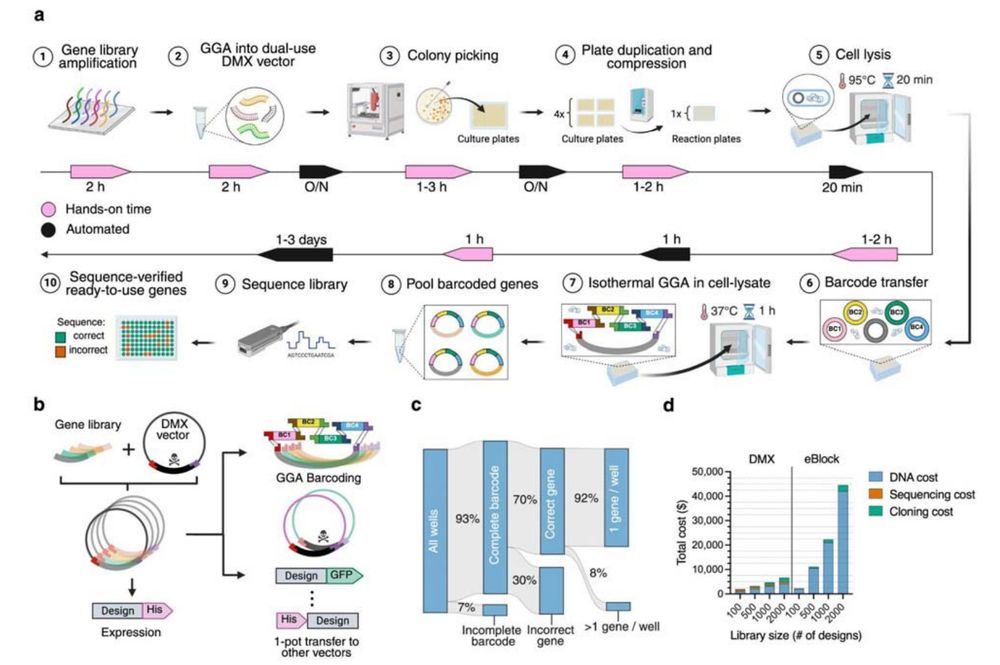
- A rapid, scalable, pipeline for producing and characterizing proteins
- A demultiplexing protocol for converting oligopools to clonal constructs
Jason Qian @lfmilles.bsky.social Basile Wicky
- A rapid, scalable, pipeline for producing and characterizing proteins
- A demultiplexing protocol for converting oligopools to clonal constructs
Jason Qian @lfmilles.bsky.social Basile Wicky
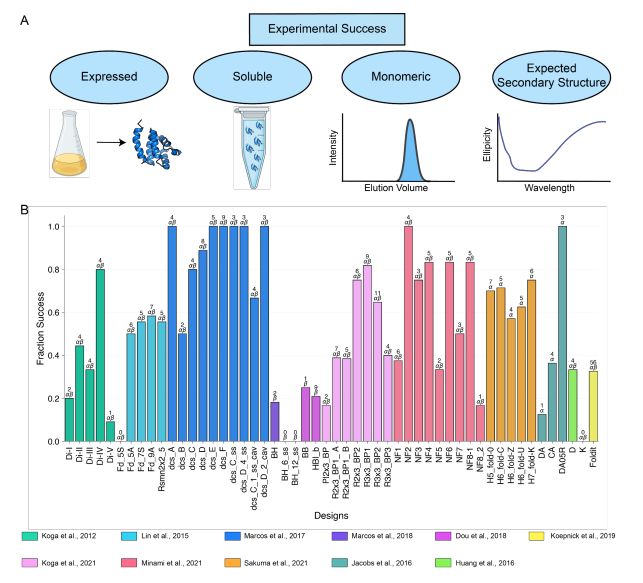
- deep learning structural metrics only weakly predict success
- The score distribution is different for different types of structures
@grocklin.bsky.social



- deep learning structural metrics only weakly predict success
- The score distribution is different for different types of structures
@grocklin.bsky.social
@yoakiyama.bsky.social Zhidian Zhang @milot.bsky.social @martinsteinegger.bsky.social @sokrypton.org



@yoakiyama.bsky.social Zhidian Zhang @milot.bsky.social @martinsteinegger.bsky.social @sokrypton.org
- predict protein-protein interactions, operon structure, and protein function
- infer phenotypic traits
- design synthetic genomes with desired properties
@macwiatrak.bsky.social @mariabrbic.bsky.social
@andresfloto.bsky.social
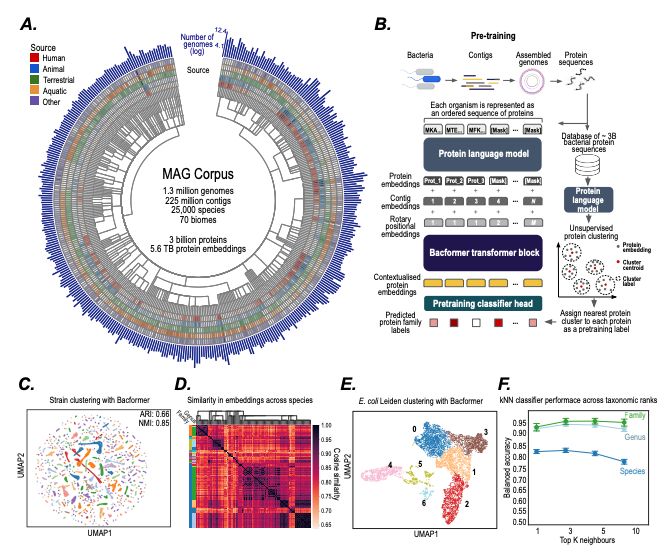
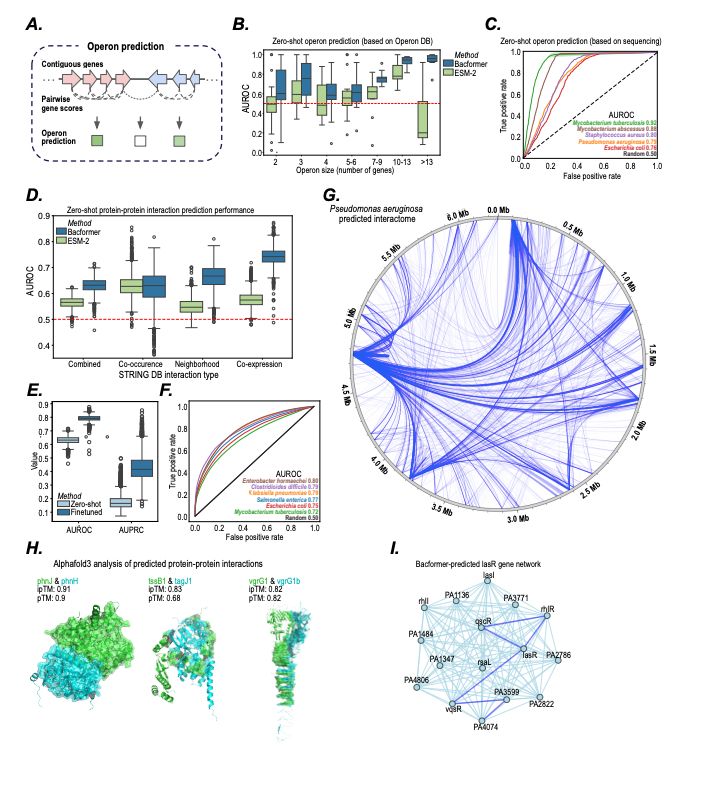
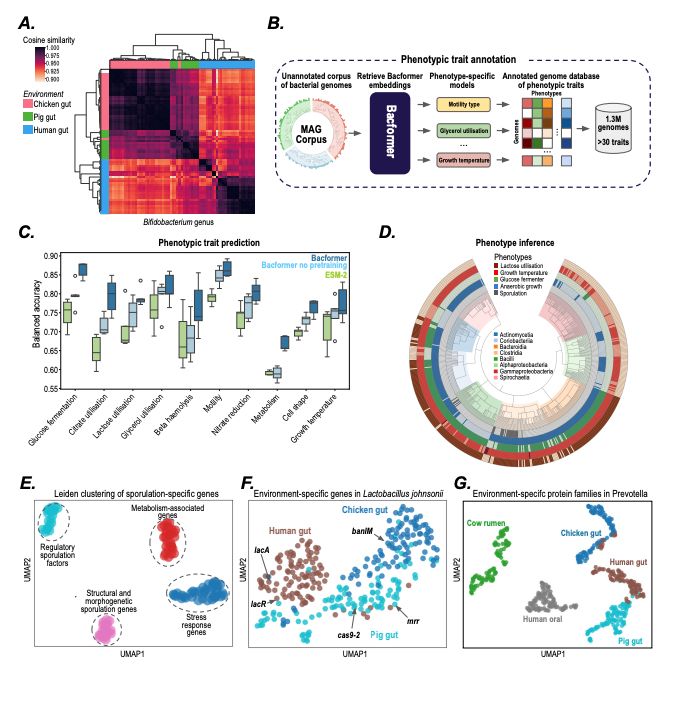
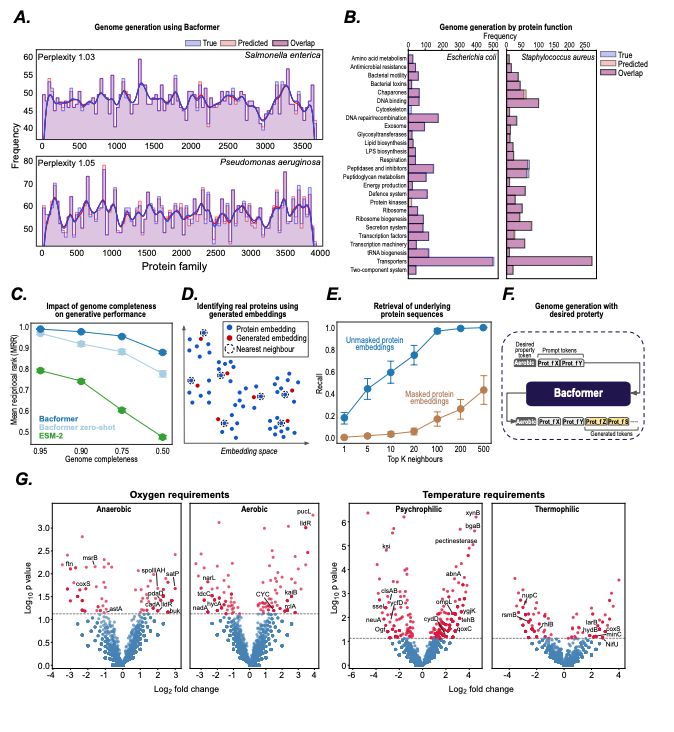
- predict protein-protein interactions, operon structure, and protein function
- infer phenotypic traits
- design synthetic genomes with desired properties
@macwiatrak.bsky.social @mariabrbic.bsky.social
@andresfloto.bsky.social




Inspired by that Atlas, today we are releasing the Dayhoff Atlas of protein sequence data and protein language models.
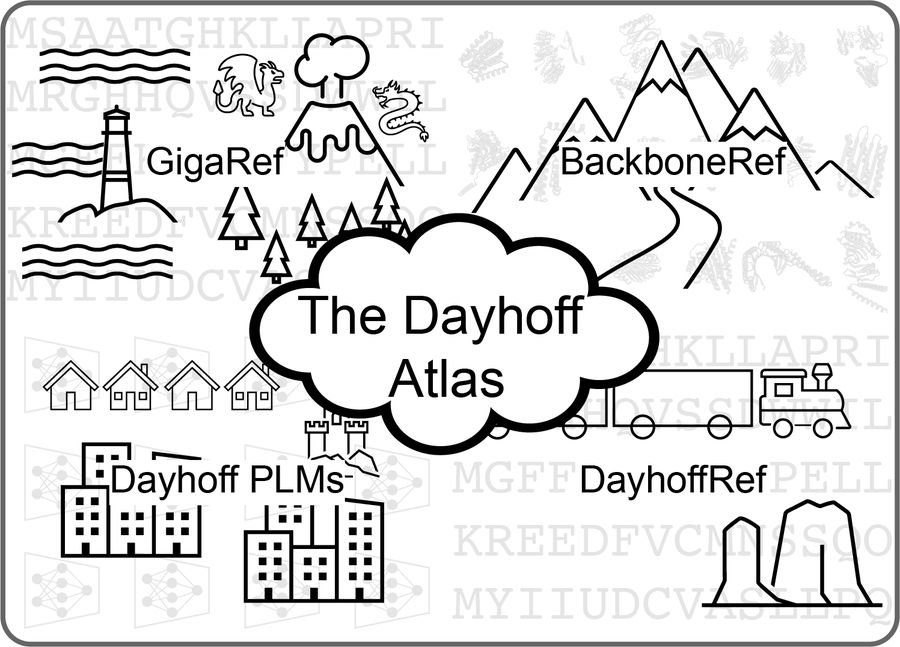
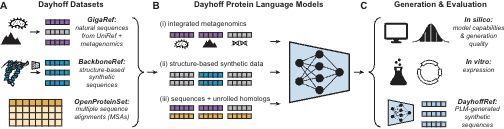
Inspired by that Atlas, today we are releasing the Dayhoff Atlas of protein sequence data and protein language models.
@kdidi.bsky.social @machine.learning.bio @karstenkreis.bsky.social @arashv.bsky.social
arxiv.org/html/2507.09...
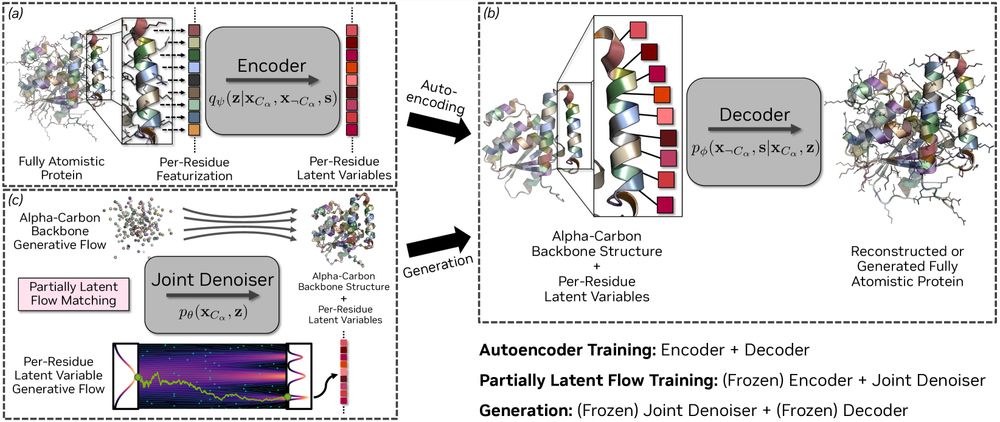
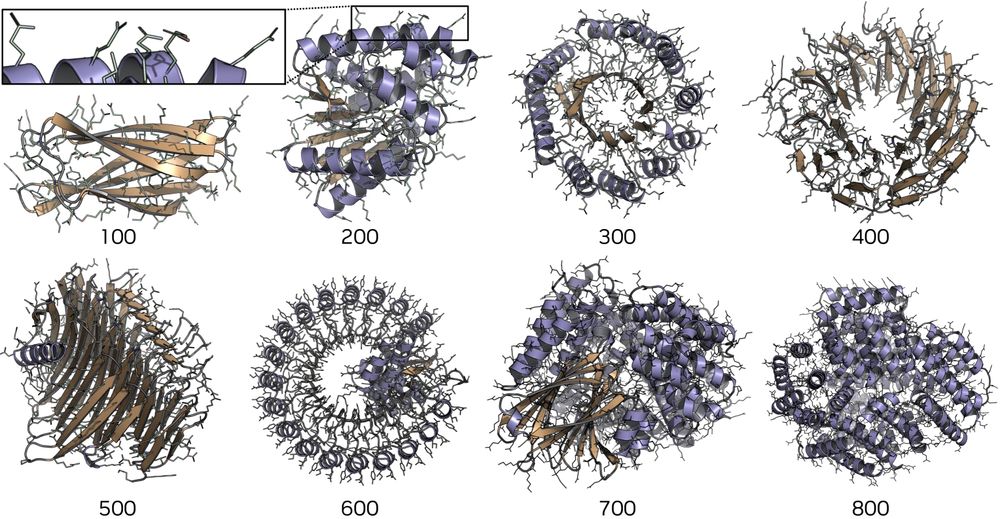

@kdidi.bsky.social @machine.learning.bio @karstenkreis.bsky.social @arashv.bsky.social
arxiv.org/html/2507.09...
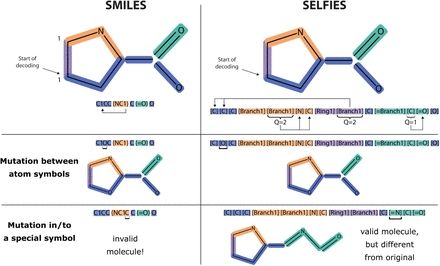
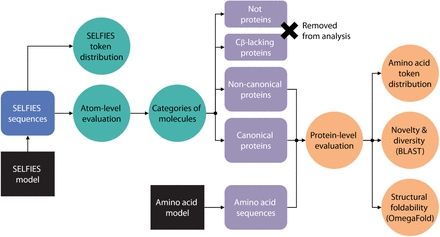

@lynnkamerlin.bsky.social
www.nature.com/articles/s41...
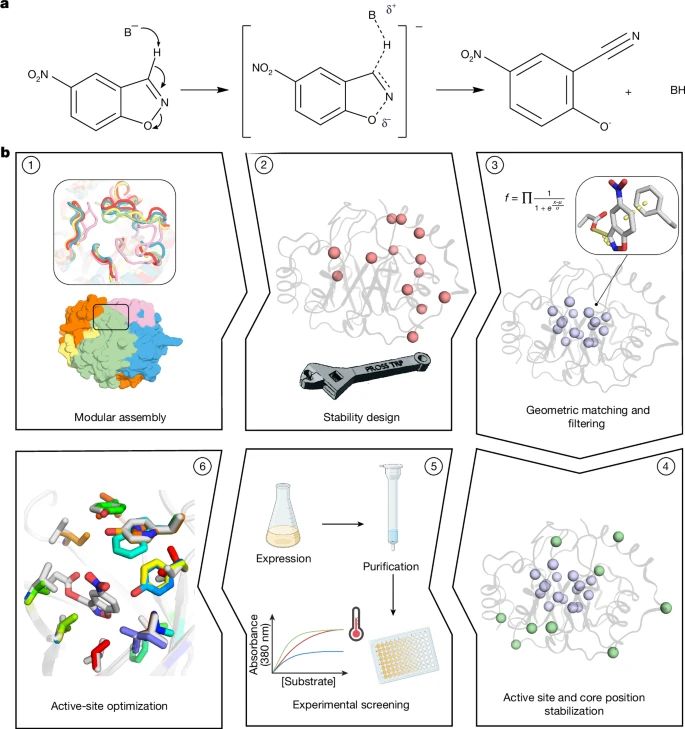
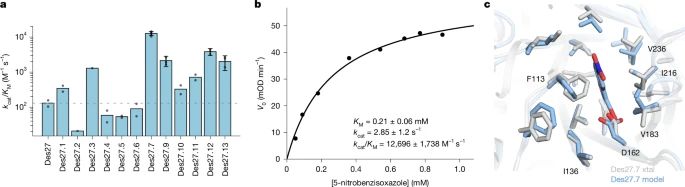
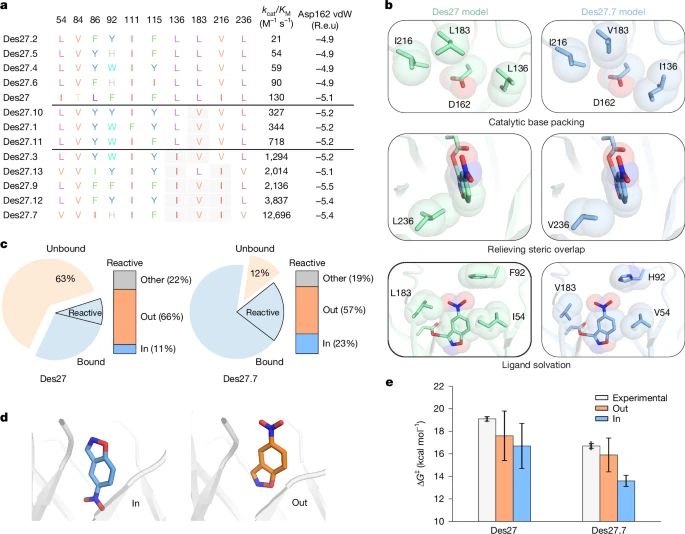
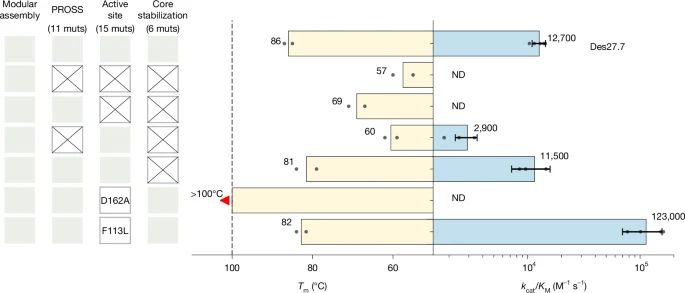
@lynnkamerlin.bsky.social
www.nature.com/articles/s41...

