
🌊 Ph.D. in Marine Science @UTMSI, University of Texas at Austin | Dr. Brett Baker’s Lab
🧬 Exploring Asgard archaea, deep-sea ecosystems, and the evolution of complex life.

We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
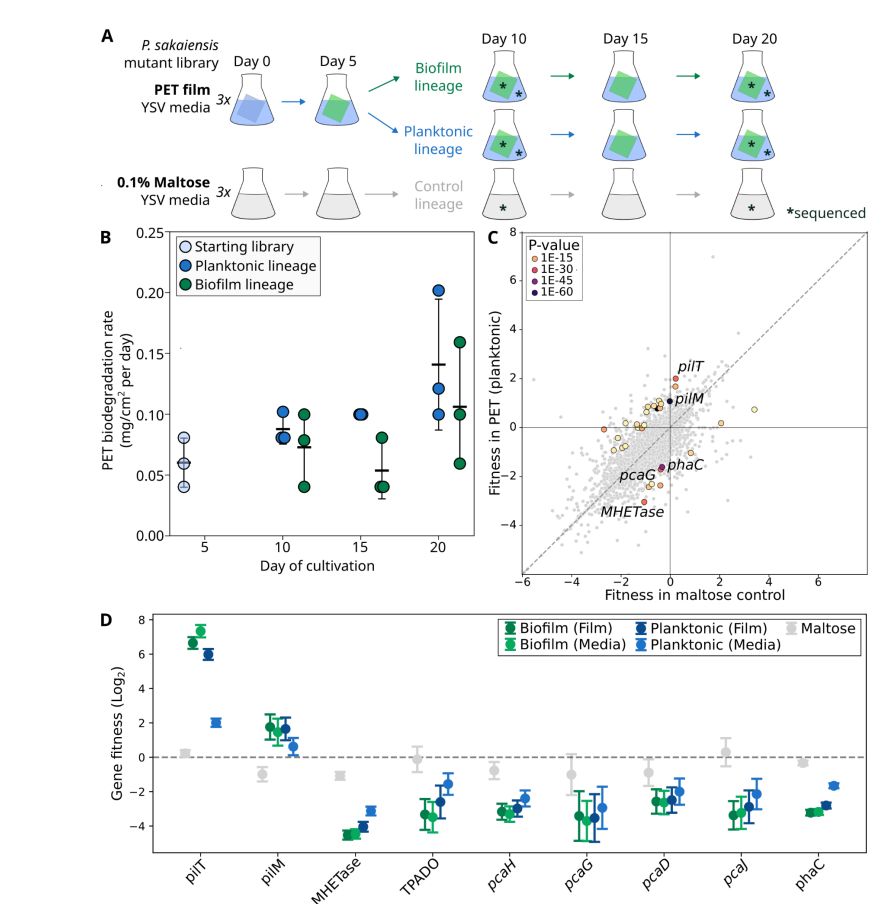
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
Excited to be a part of this project identifying a broad diversity of deep-ocean plastic-degrading enzymes (PETases/BHETases).
doi.org/10.1093/isme...
Excited to be a part of this project identifying a broad diversity of deep-ocean plastic-degrading enzymes (PETases/BHETases).
doi.org/10.1093/isme...
We obtained lots of thermal stable plastic degrading enzymes from the deep sea (Guaymas Basin, Gulf of California)

We obtained lots of thermal stable plastic degrading enzymes from the deep sea (Guaymas Basin, Gulf of California)

We present a foundational genomic resource of human gut microbiome viruses. It delivers high-quality, deeply curated data spanning taxonomy, predicted hosts, structures, and functions, providing a reference for gut virome research. (1/8)
www.biorxiv.org/content/10.1...

We present a foundational genomic resource of human gut microbiome viruses. It delivers high-quality, deeply curated data spanning taxonomy, predicted hosts, structures, and functions, providing a reference for gut virome research. (1/8)
www.biorxiv.org/content/10.1...
Here’s the story of how we discovered the Metis defense system 👇
www.biorxiv.org/content/10.1...

Here’s the story of how we discovered the Metis defense system 👇
www.biorxiv.org/content/10.1...
Congrats Romain and team!!
Congrats Romain and team!!
@mkrupovic.bsky.social @deemteam.bsky.social @anagtz.bsky.social
www.nature.com/articles/s41...

@mkrupovic.bsky.social @deemteam.bsky.social @anagtz.bsky.social
www.nature.com/articles/s41...
We challenge the long-standing view that peptidoglycan alone protects cells from bursting.
Our study shows that the periplasm — enclosed by OM–PG connections — acts as a pressure buffer essential for osmoprotection in Gram-negative bacteria.
📄 www.nature.com/articles/s41...
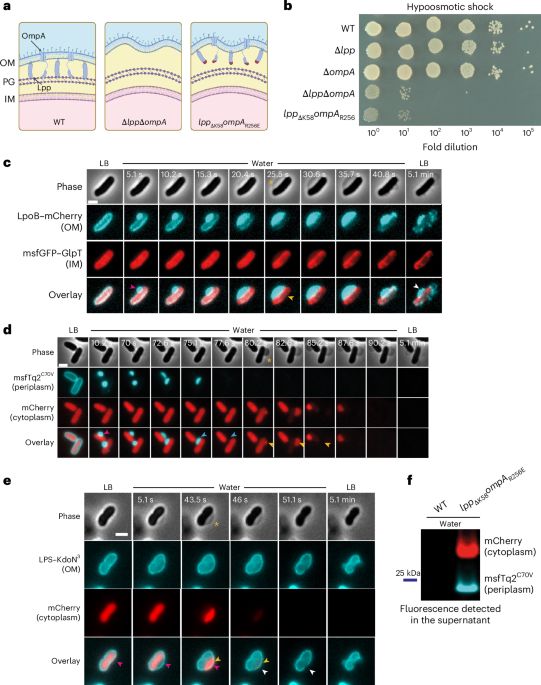
We challenge the long-standing view that peptidoglycan alone protects cells from bursting.
Our study shows that the periplasm — enclosed by OM–PG connections — acts as a pressure buffer essential for osmoprotection in Gram-negative bacteria.
📄 www.nature.com/articles/s41...
www.nature.com/articles/s41...
www.nature.com/articles/s41...
go.nature.com/4mvYGmd

go.nature.com/4mvYGmd
Palaeocampa is an exceptional lobopodian - it lived in rivers and lakes, bristled with thousands of poisonous spines, and more. 🧵
Open access: nature.com/articles/s42...

Palaeocampa is an exceptional lobopodian - it lived in rivers and lakes, bristled with thousands of poisonous spines, and more. 🧵
Open access: nature.com/articles/s42...
My first research paper I've been involved with is out now as a preprint 🦠🥳🦠
#archaea #microsky #PhD
Big group effort with a lot of people involved
#archaeasky #archaea
www.biorxiv.org/content/10.1...

www.nature.com/articles/s41...
Open access link: rdcu.be/exnOc
Many years in the making, great collaboration with @archaellum.bsky.social & @tunglejic.bsky.social
Thanks @ukri.org BBSRC & @leverhulme.ac.uk for funding, reviewers & editor!! 🙏
#microsky #archaeasky
www.nature.com/articles/s41...
Open access link: rdcu.be/exnOc
Many years in the making, great collaboration with @archaellum.bsky.social & @tunglejic.bsky.social
Thanks @ukri.org BBSRC & @leverhulme.ac.uk for funding, reviewers & editor!! 🙏
#microsky #archaeasky
rdcu.be/euila
Our main results stay the same: S. islandicus follows a cell cycle akin to that of some eukaryotes, like the budding yeast, with cellular processes cycling similarly.
old🧵cited below👇
rdcu.be/euila
Our main results stay the same: S. islandicus follows a cell cycle akin to that of some eukaryotes, like the budding yeast, with cellular processes cycling similarly.
old🧵cited below👇
🧵 New preprint out!
WitChi: a fast, open-source Python tool to detect, quantify & prune compositional bias in MSAs.
Lightweight, tree-free, scalable to 5k+ taxa... so we applied it to the GTDB archaea MSA.
#ArchaeaSky #MEvoSky #MicroSky
🔗 doi.org/10.1101/2025...
💻 github.com/stephkoest/w...

🧵 New preprint out!
WitChi: a fast, open-source Python tool to detect, quantify & prune compositional bias in MSAs.
Lightweight, tree-free, scalable to 5k+ taxa... so we applied it to the GTDB archaea MSA.
#ArchaeaSky #MEvoSky #MicroSky
🔗 doi.org/10.1101/2025...
💻 github.com/stephkoest/w...
We tested it on the GTDB r220 archaeal supermatrix (5,869 taxa & 10,101 cols) removing 55% of sites in <2h.
The phylogeny showed several interesting groupings with overall improved branch support:
#phylogenetics #ArchaeaSky #MSA #opensource #MEvoSky #MicroSky

We tested it on the GTDB r220 archaeal supermatrix (5,869 taxa & 10,101 cols) removing 55% of sites in <2h.
The phylogeny showed several interesting groupings with overall improved branch support:
#phylogenetics #ArchaeaSky #MSA #opensource #MEvoSky #MicroSky
🏆 Winner: Dr Hannah Brown @hannjbrown.bsky.social
🏆 Runner-Up: Dr Binod Rayamajhee
We’re proud to support the next generation of microbiology leaders and can’t wait to see where their research takes them!






