Heidelberg University, codirector DREAM challenges. For group's activities, see @saezlab.bsky.social
reflecting from my perspective as patient-scientist (www.nature.com/articles/s41...) & sharing work of @saezlab.bsky.social sp. within www.iganatlas.org
reflecting from my perspective as patient-scientist (www.nature.com/articles/s41...) & sharing work of @saezlab.bsky.social sp. within www.iganatlas.org
➡️ www.nature.com/articles/s41...
A case study shows MCP servers working together - combining BioContextAI Knowledgebase with our omnipath omnipathdb.org MCP (work in progress) to showcase interoperability. Looking forward to see how the ecosystem evolves!
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
➡️ www.nature.com/articles/s41...
doi.org/10.1101/2024...

The new abstract submission deadline for #EMBLCardioBiology is 10 November 👉 https://s.embl.org/ncb26-01-bl
👉 Join experts pushing the frontiers of heart and vascular science
🫀 Share your work and shape the future of cardiobiology
📜 Submit your abstract today!

The new abstract submission deadline for #EMBLCardioBiology is 10 November 👉 https://s.embl.org/ncb26-01-bl
👉 Join experts pushing the frontiers of heart and vascular science
🫀 Share your work and shape the future of cardiobiology
📜 Submit your abstract today!
It was really interesting to write a short manuscript mixing method presentation & broader insights on data preprocessing. 📝
Thank you to my co-supervisors @cantinilab.bsky.social & @juliosaezrod.bsky.social for their feedback on this side work! 🫶
5/5
It was really interesting to write a short manuscript mixing method presentation & broader insights on data preprocessing. 📝
Thank you to my co-supervisors @cantinilab.bsky.social & @juliosaezrod.bsky.social for their feedback on this side work! 🫶
5/5
Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️

Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️
Details & apply by 13/10/25: tinyurl.com/4shdw8dk

Details & apply by 13/10/25: tinyurl.com/4shdw8dk
📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️

📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️
We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape

We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io

🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io
Jess’s group takes a multi-disciplinary approach to identify and characterise chemical hazards to humans and ecosystems, using cell profiling data, machine learning, and integrative data analysis.
www.ebi.ac.uk/about/news/p...
🖥️🧬

Jess’s group takes a multi-disciplinary approach to identify and characterise chemical hazards to humans and ecosystems, using cell profiling data, machine learning, and integrative data analysis.
www.ebi.ac.uk/about/news/p...
🖥️🧬
Find out how Tim’s group is exploring using large-scale single-cell and spatial data to trace cell lineages, understand cancer origins, and uncover how mutations drive disease.
www.ebi.ac.uk/about/news/p...
🖥️🧬

Find out how Tim’s group is exploring using large-scale single-cell and spatial data to trace cell lineages, understand cancer origins, and uncover how mutations drive disease.
www.ebi.ac.uk/about/news/p...
🖥️🧬
At EMBL, we train young scientists to become skilled and creative future leaders in academia and other sectors. Join us and get a head start on your career in life sciences!
bit.ly/47mmiFF



🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️

🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️
A case study shows MCP servers working together - combining BioContextAI Knowledgebase with our omnipath omnipathdb.org MCP (work in progress) to showcase interoperability. Looking forward to see how the ecosystem evolves!
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
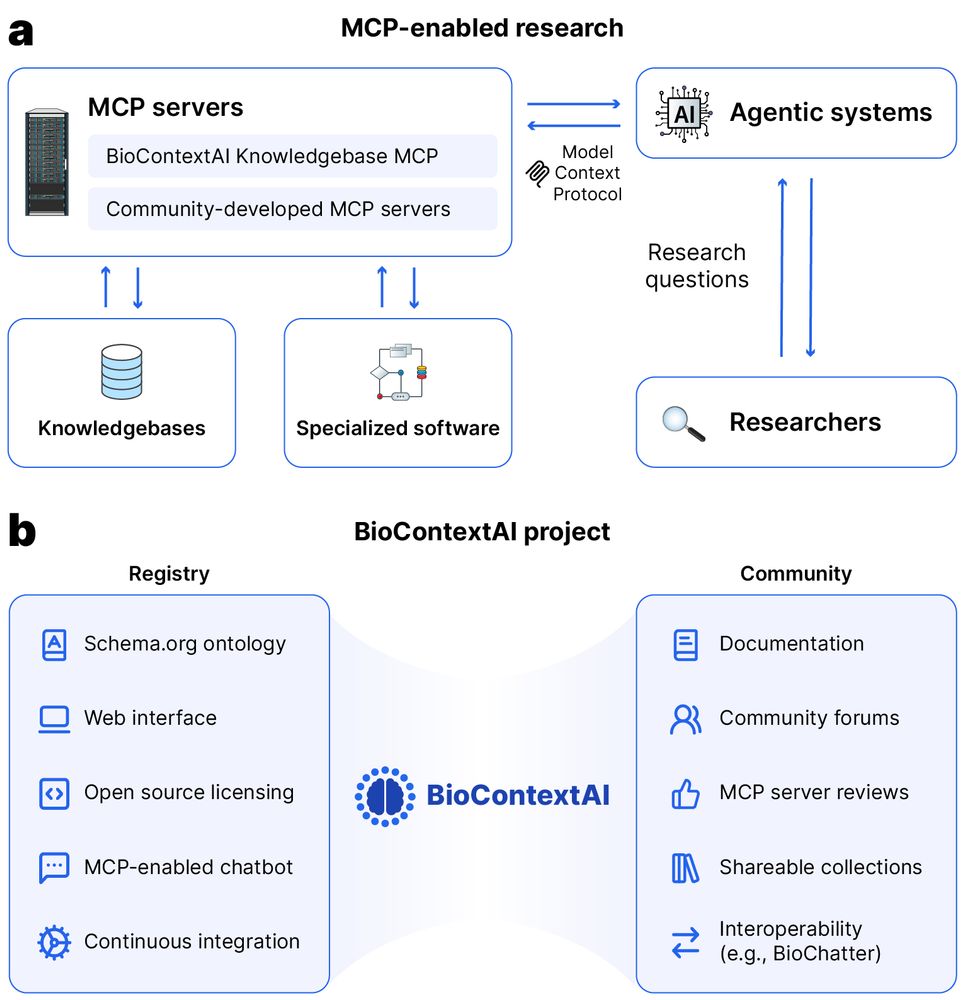
A case study shows MCP servers working together - combining BioContextAI Knowledgebase with our omnipath omnipathdb.org MCP (work in progress) to showcase interoperability. Looking forward to see how the ecosystem evolves!
1️⃣ Multicellular molecular characterisation of inflammatory bowel disease
6️⃣ Designing Efficient Single-Cell Perturbation Experiments with Lab-in-the-Loop AI Agents
📅 Application deadline is 30th September ❗
ESPOD builds on the collaborative relationship between EMBL-EBI and the @sangerinstitute.bsky.social, offering projects that combine experimental and computational approaches.
www.ebi.ac.uk/research/pos...
🧬🖥️🔬#postdocjob

1️⃣ Multicellular molecular characterisation of inflammatory bowel disease
6️⃣ Designing Efficient Single-Cell Perturbation Experiments with Lab-in-the-Loop AI Agents
📅 Application deadline is 30th September ❗
Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...

Or would you like to find a way to use biomedical tools using natural language?
Then check out biocontext.ai, now out on bioRxiv: www.biorxiv.org/content/10.1...
www.embl.org/training/ari...
www.embl.org/training/ari...
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
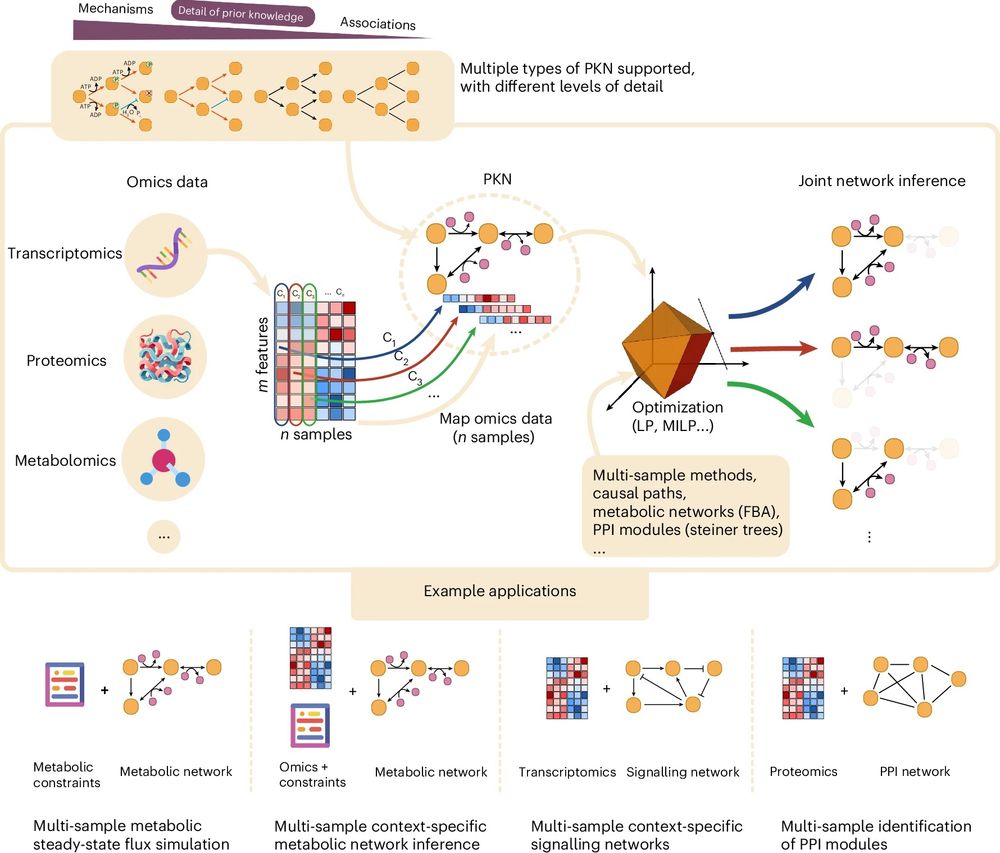
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
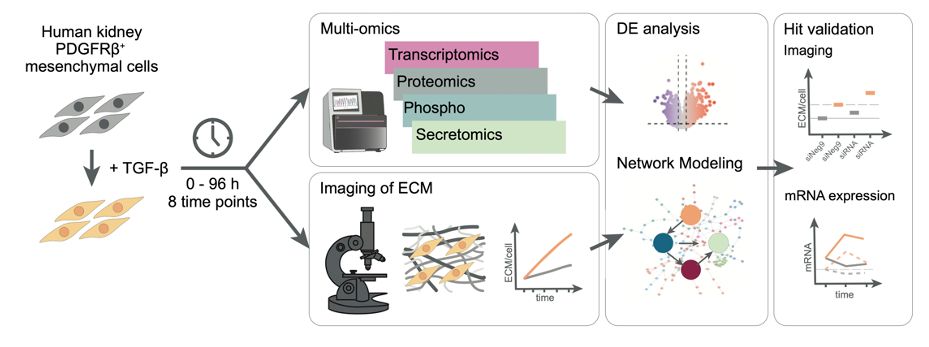


This release includes a streamlined API aligned with scverse standards, updated vignettes, and a new one on pseudotime enrichment analysis 👇
decoupler.readthedocs.io/en/latest/
This release includes a streamlined API aligned with scverse standards, updated vignettes, and a new one on pseudotime enrichment analysis 👇
decoupler.readthedocs.io/en/latest/

