---
#proteomics #prot-paper

---
#proteomics #prot-paper
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...

AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...
📄 www.nature.com/articles/s41...
#LoveVirology #TeamMassSpec
📄 www.nature.com/articles/s41...
#LoveVirology #TeamMassSpec
www.nature.com/articles/s41...

www.nature.com/articles/s41...
#Proteomics #TeamMassSpec
www.embopress.org/doi/full/10....

#Proteomics #TeamMassSpec
www.embopress.org/doi/full/10....
Software: github.com/KosinskiLab/...
Preprint: biorxiv.org/content/10.1...


Software: github.com/KosinskiLab/...
Preprint: biorxiv.org/content/10.1...
#XLMS #Massspec #Proteomics
www.nature.com/articles/s41...

#XLMS #Massspec #Proteomics
www.nature.com/articles/s41...

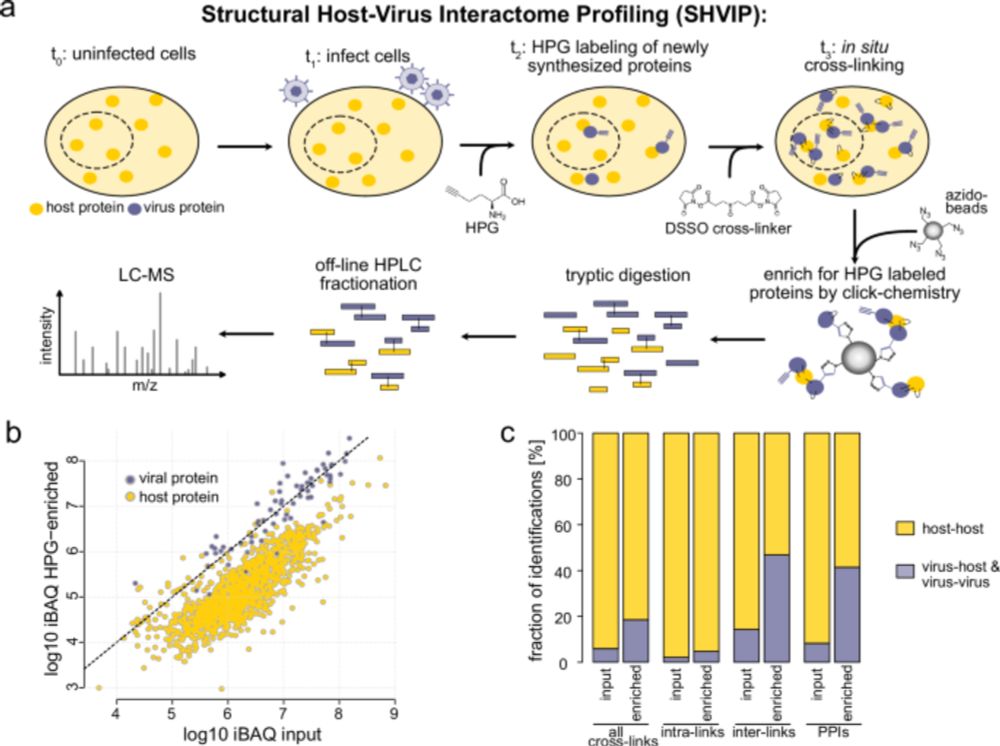
In our new preprint, we achieve higher coverage virus-host crosslinks than traditional interaction proteomics approaches by specifically labeling viral proteins, and can characterize new interaction sites in disordered regions via AF-multimer.
In our new preprint, we achieve higher coverage virus-host crosslinks than traditional interaction proteomics approaches by specifically labeling viral proteins, and can characterize new interaction sites in disordered regions via AF-multimer.

