
Screenshotting the image here from a 1994 book on nuclear physics, for others who may be interested:
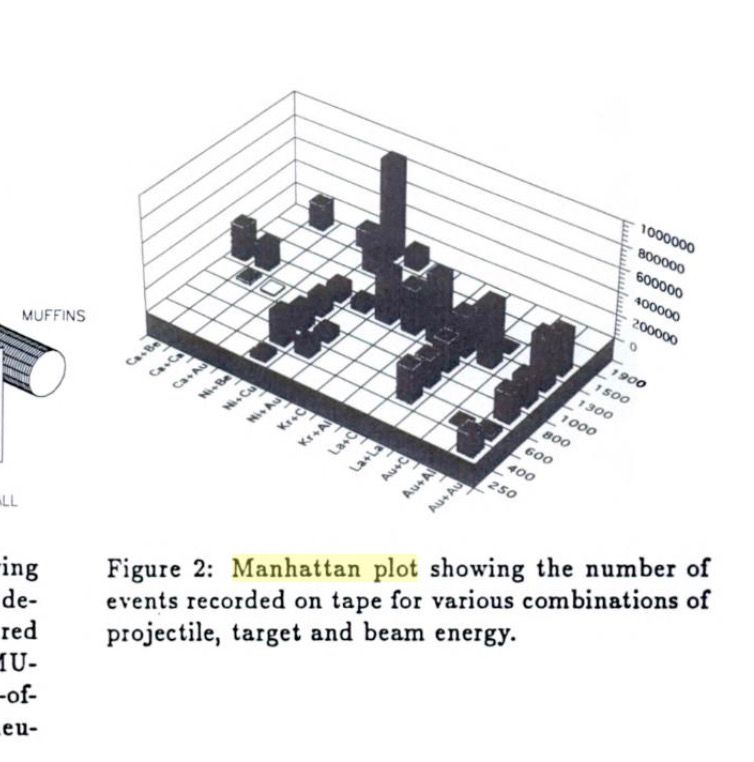
Screenshotting the image here from a 1994 book on nuclear physics, for others who may be interested:

Followup, does anyone know who made the original version so that I can credit them?

Followup, does anyone know who made the original version so that I can credit them?
(I know many of you are waiting for the GWAS chapters of my free online textbook on human genetics--they are coming, but slowly!)

(I know many of you are waiting for the GWAS chapters of my free online textbook on human genetics--they are coming, but slowly!)

We inferred cell cycle stage in all the Perturb-seq cells to check our model and got something curious: regulators had the expected direction of effect on the fraction of cells in each stage, but program genes didn't really change the fractions.

We inferred cell cycle stage in all the Perturb-seq cells to check our model and got something curious: regulators had the expected direction of effect on the fraction of cells in each stage, but program genes didn't really change the fractions.
Prediction: R_a genes should have much stronger effects on MCH than R_b genes. This is in fact correct!
Even being able to test this is a huge win for quantitative effect sizes

Prediction: R_a genes should have much stronger effects on MCH than R_b genes. This is in fact correct!
Even being able to test this is a huge win for quantitative effect sizes


We noticed that the regulator genes are largely shared across 3 critical programs: G2/M and S phase, and autophagy.
Here are the Perturb-seq effects of G2/M regulators on S and autophagy. The phases of cell cycle are oppositely regulated, and both repress autophagy.

We noticed that the regulator genes are largely shared across 3 critical programs: G2/M and S phase, and autophagy.
Here are the Perturb-seq effects of G2/M regulators on S and autophagy. The phases of cell cycle are oppositely regulated, and both repress autophagy.
For example, for S phase the program genes repress MCH, but the regulators activate MCH. How could we explain this?
[Remember, we wouldn't have noticed this without the directional information implicit in LoFs!]

For example, for S phase the program genes repress MCH, but the regulators activate MCH. How could we explain this?
[Remember, we wouldn't have noticed this without the directional information implicit in LoFs!]

For each program we asked if the Program or the Regulators show a significant DIRECTIONAL effect on each trait.

For each program we asked if the Program or the Regulators show a significant DIRECTIONAL effect on each trait.
As expected, positive regulators of HBA1 (knockdown reduces expression) are also positive regulators of MCH (LoFs lower MCH)

As expected, positive regulators of HBA1 (knockdown reduces expression) are also positive regulators of MCH (LoFs lower MCH)
eg here's a volcano plot of LoF tests for hemoglobin (MCH) in UKB. As you would expect the biggest negative hits include hemoglobin components: HBB, HBA1, HBA2.

eg here's a volcano plot of LoF tests for hemoglobin (MCH) in UKB. As you would expect the biggest negative hits include hemoglobin components: HBB, HBA1, HBA2.


This highlights programs including hemoglobin synthesis, cell cycle, autophagy, and progenitor maintenance.

This highlights programs including hemoglobin synthesis, cell cycle, autophagy, and progenitor maintenance.
Next we test for DIRECTIONAL relationships between LoF gene effects and regulators and programs.

Next we test for DIRECTIONAL relationships between LoF gene effects and regulators and programs.
LoFs help for 2 reasons: they give us stronger enrichment in regulators, and they are inherently DIRECTIONAL, information we use later.

LoFs help for 2 reasons: they give us stronger enrichment in regulators, and they are inherently DIRECTIONAL, information we use later.
Using directional information from LoF data we see a strong signal that this is true. Game on!

Using directional information from LoF data we see a strong signal that this is true. Game on!
But this is still a very limited picture. eg, WHICH aspects of cell cycle matter? WHY? And does c.c. increase or decrease MCH?


But this is still a very limited picture. eg, WHICH aspects of cell cycle matter? WHY? And does c.c. increase or decrease MCH?

I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵
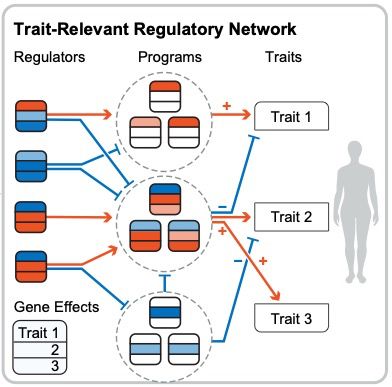
I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵

