
Source: johnhw.github.io/uma...

Source: johnhw.github.io/uma...


We present ideas on how we might end up training models from atoms to organs by using transformers to compress 🔺 🔻 data into tokens used by larger scale models
We present ideas on how we might end up training models from atoms to organs by using transformers to compress 🔺 🔻 data into tokens used by larger scale models
www.biorxiv.org/cont...
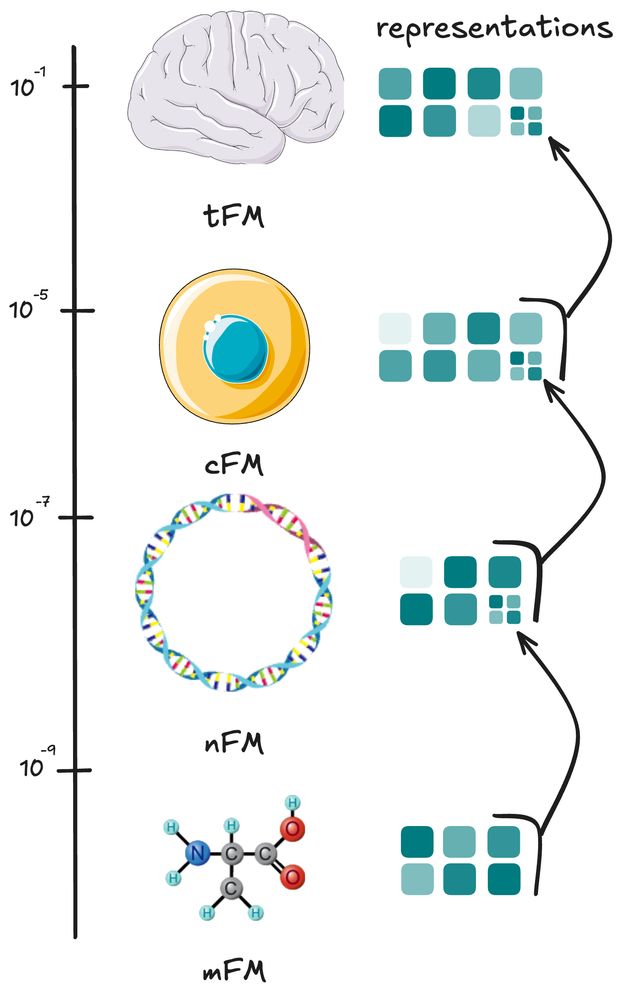
Foundation Models are being trained from atoms to molecules ⚛️, molecule chains 🧬, entire cells 🦠, and even groups of cell across tissue slices 🫁

www.biorxiv.org/cont...
Foundation Models are being trained from atoms to molecules ⚛️, molecule chains 🧬, entire cells 🦠, and even groups of cell across tissue slices 🫁
I also put together a short video recap (in French) for those curious: youtu.be/fc8L8Dn_7tw...
1/2

I also put together a short video recap (in French) for those curious: youtu.be/fc8L8Dn_7tw...
1/2
I also put together a short video recap (in French) for those curious: youtu.be/fc8L8Dn_7tw...
1/2

I also put together a short video recap (in French) for those curious: youtu.be/fc8L8Dn_7tw...
1/2
6/6

6/6
5/6

5/6
4/6

4/6
3/6

3/6
2/6

2/6



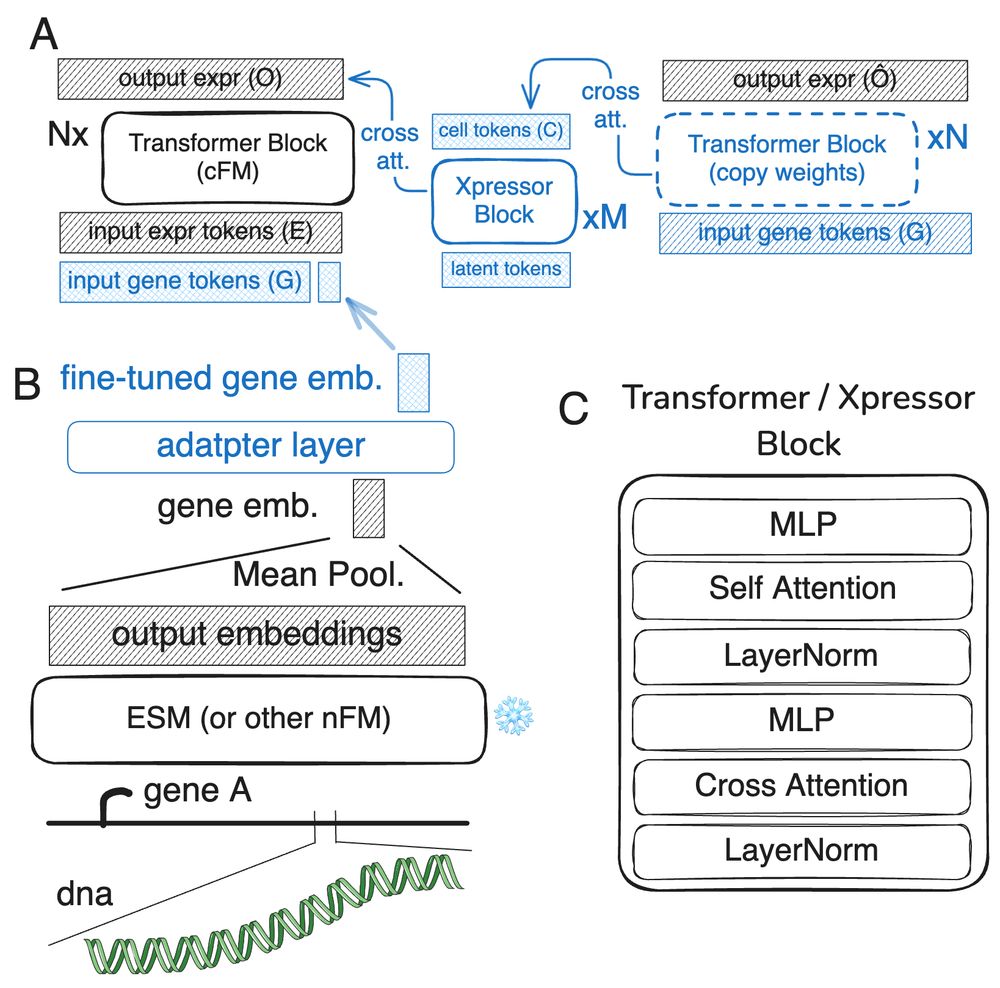
Transformer use embeddings of element they look at. each can be produced by the previous scale transformer model. going from molecules to whole tissues!

Transformer use embeddings of element they look at. each can be produced by the previous scale transformer model. going from molecules to whole tissues!

We also propose a new hierarchical classification method to work with the rich hierarchical ontologies used to label cells in cellxgene.


We also propose a new hierarchical classification method to work with the rich hierarchical ontologies used to label cells in cellxgene.
scPRINT is a transformer model trained on 50M cells 🦠 from the cellxgene database, it has novel expression encoding and decoding schemes and new pre-training methodologies 🤖.
www.biorxiv.org/content/10.1...

scPRINT is a transformer model trained on 50M cells 🦠 from the cellxgene database, it has novel expression encoding and decoding schemes and new pre-training methodologies 🤖.
www.biorxiv.org/content/10.1...


