
Have been working on motor protein, microtubule, and cilia.
Recently also working with membrane proteins.
Associate Editorial Board member of Cytoskeleton Journal.
Lab website: https://tinyurl.com/22c3eymy



7/8 🧵

7/8 🧵
6/8 🧵

6/8 🧵
5/8 🧵

5/8 🧵
4/8 🧵

4/8 🧵
3/8 🧵

3/8 🧵
Please check this amazing video taken by high-speed AFM!!!
www.nature.com/articles/s41...
Please check this amazing video taken by high-speed AFM!!!
www.nature.com/articles/s41...
blueskyroast.com

blueskyroast.com


Reported as a reviewed preprint in eLife this year.
elifesciences.org/articles/92829

Reported as a reviewed preprint in eLife this year.
elifesciences.org/articles/92829
www.nature.com/articles/s41...

www.nature.com/articles/s41...
@scholargoggler.bsky.social. I thought there were more cilia in the title, but not really. 😂
@scholargoggler.bsky.social. I thought there were more cilia in the title, but not really. 😂
www.nature.com/articles/s41...
www.nature.com/articles/s41...
www.science.org/doi/full/10....

www.science.org/doi/full/10....
www.embopress.org/doi/full/10....
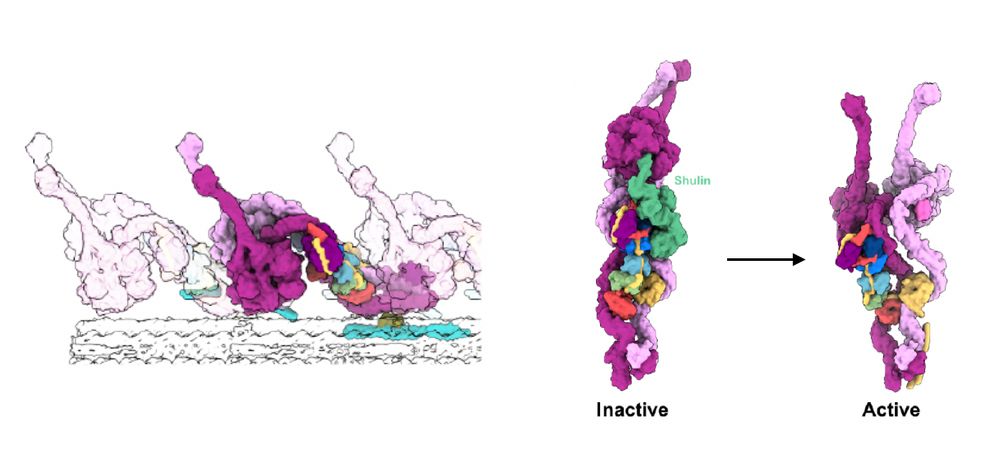
www.embopress.org/doi/full/10....

This paper was published in @naturecellbiology.bsky.social in 2014, so definitely not recent, but we proposed that dynein-1's stacked conformation is autoinhibitory state for the first time. This model is widely accepted in the field now. www.nature.com/articles/ncb...

This paper was published in @naturecellbiology.bsky.social in 2014, so definitely not recent, but we proposed that dynein-1's stacked conformation is autoinhibitory state for the first time. This model is widely accepted in the field now. www.nature.com/articles/ncb...
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
elifesciences.org/articles/52760
elifesciences.org/articles/52760
www.pnas.org/doi/abs/10.1...

www.pnas.org/doi/abs/10.1...
journals.plos.org/plosbiology/...

journals.plos.org/plosbiology/...
www.science.org/doi/full/10....

www.science.org/doi/full/10....
Cryo-EM structure of the zinc-activated channel (ZAC) in the Cys-loop receptor superfamily | PNAS
www.pnas.org/doi/10.1073/...

Cryo-EM structure of the zinc-activated channel (ZAC) in the Cys-loop receptor superfamily | PNAS
www.pnas.org/doi/10.1073/...

