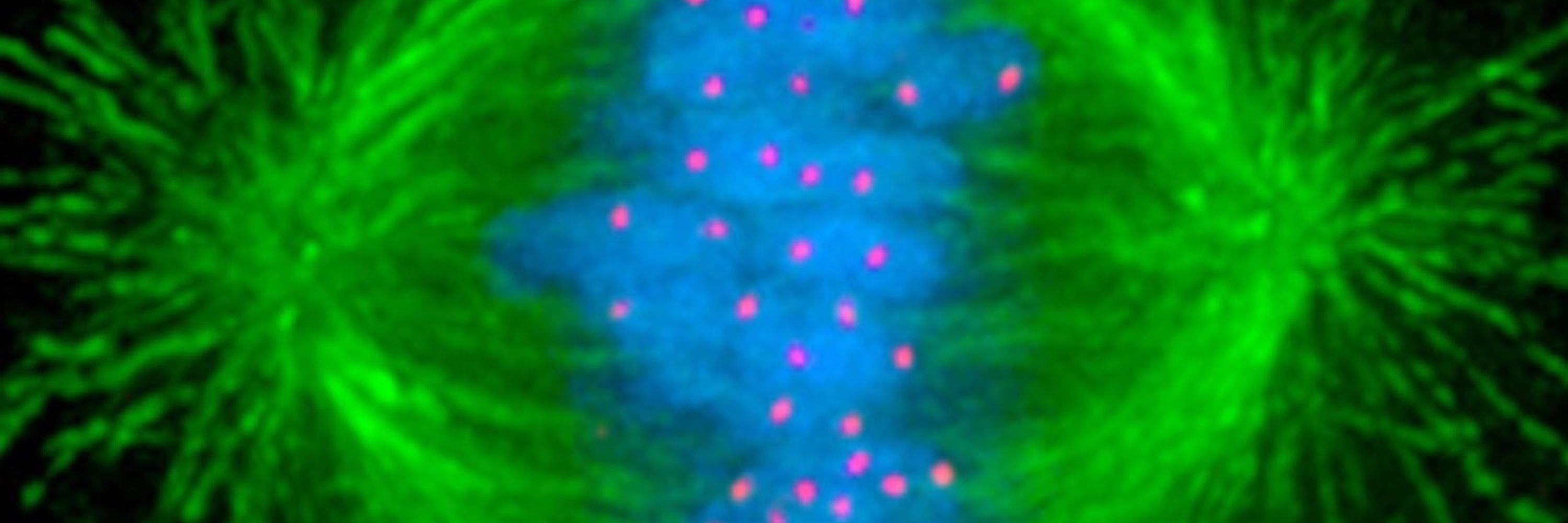
Read the paper here: lnkd.in/eTNkJhBW
Read the paper here: lnkd.in/eTNkJhBW
As a leader, I have also appreciated it when others did the same to (for) me.
/fin

As a leader, I have also appreciated it when others did the same to (for) me.
/fin
www.nature.com/articles/s41...

www.nature.com/articles/s41...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

