Designing embedded systems for bioinformatics applications.
Now the pod5 (while indeed better than fast5) is still behind slow5 in this regards
academic.oup.com/gigascience/...

Now the pod5 (while indeed better than fast5) is still behind slow5 in this regards
academic.oup.com/gigascience/...
The output produced was wrong - A simple program that sums up the signal values.
I just checked release logs of pod5 - 0.0.20 "Fix bug reading data via C API". So it could be a bug that got introduced in before.
The output produced was wrong - A simple program that sums up the signal values.
I just checked release logs of pod5 - 0.0.20 "Fix bug reading data via C API". So it could be a bug that got introduced in before.
It is very interesting that they have skipped those questions 🤣

It is very interesting that they have skipped those questions 🤣
We plan to try this out with adaptive sampling for metagenomes, like we did in cornetto with hifiasm [https://doi.org/10.1101/2025.03.31.646505] for genome assemblies.
We plan to try this out with adaptive sampling for metagenomes, like we did in cornetto with hifiasm [https://doi.org/10.1101/2025.03.31.646505] for genome assemblies.
I am glad that some time ago, I disabled this Gemini summary from my search results when it was pushed out. Seems I should better keep it that way😂
I am glad that some time ago, I disabled this Gemini summary from my search results when it was pushed out. Seems I should better keep it that way😂
Thank you very much for the suggestion
Thank you very much for the suggestion
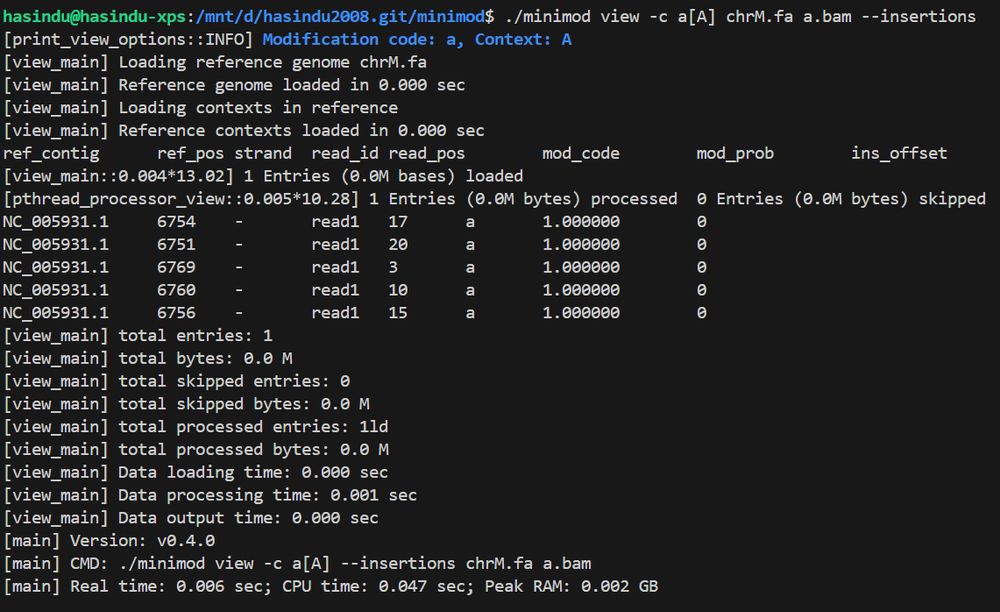
6754
6751
6769
6760
6756
Which seem to match the expected, assuming you are using 1-based coordinates
6754
6751
6769
6760
6756
Which seem to match the expected, assuming you are using 1-based coordinates

