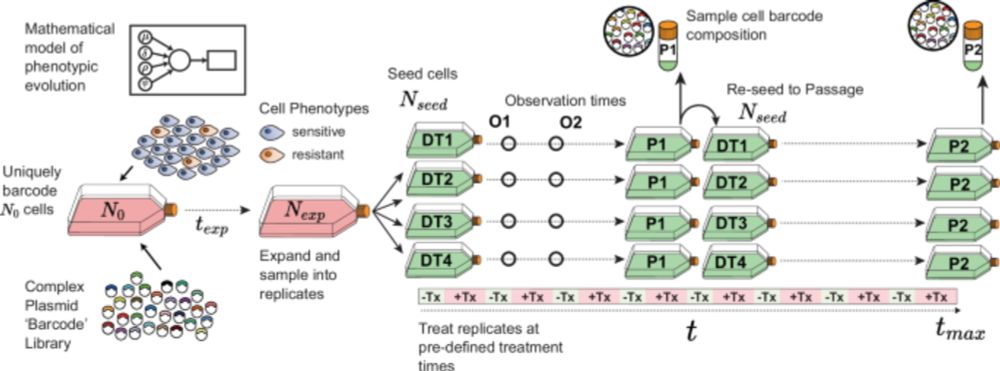



www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

rdcu.be/eFrrc

rdcu.be/eFrrc
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
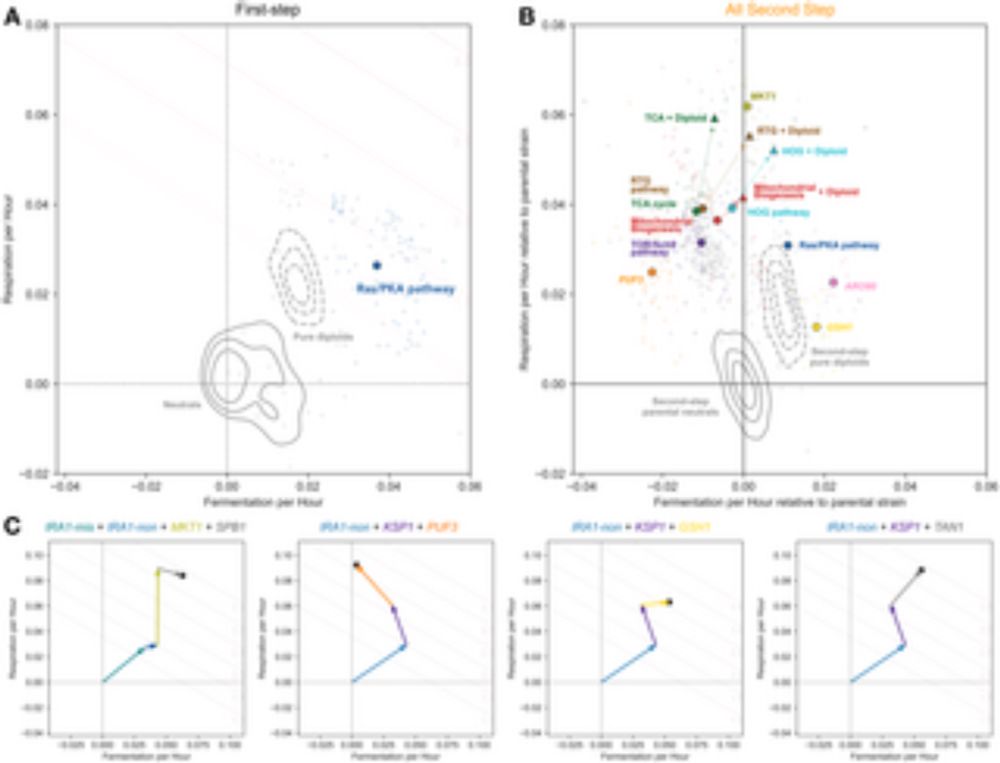
recruit.ap.uci.edu/JPF09601

recruit.ap.uci.edu/JPF09601

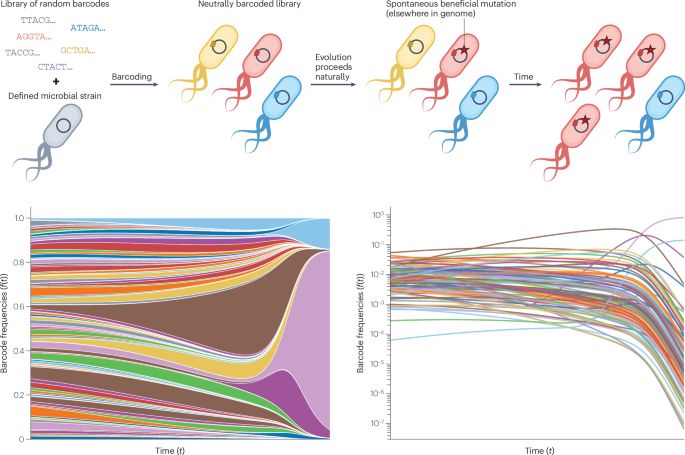
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
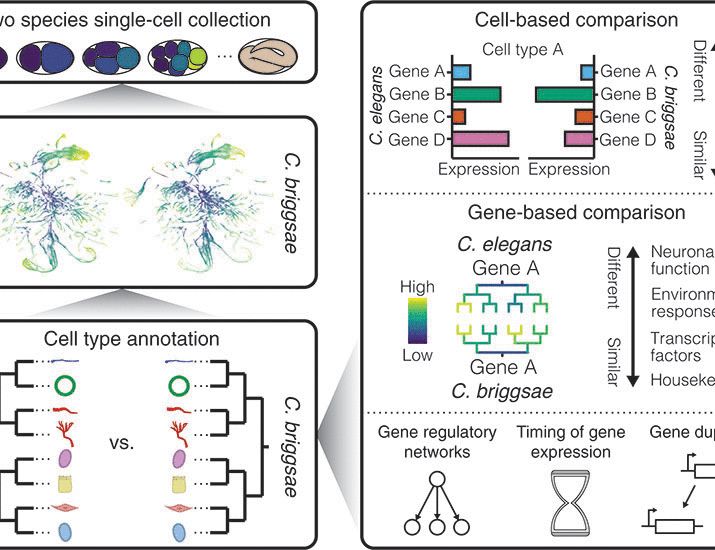
👉 Early cancer detection
👉 Methylation drivers
👉 Epimutation rates
👉 CpG lineage tracing
www.biorxiv.org/content/10.1...
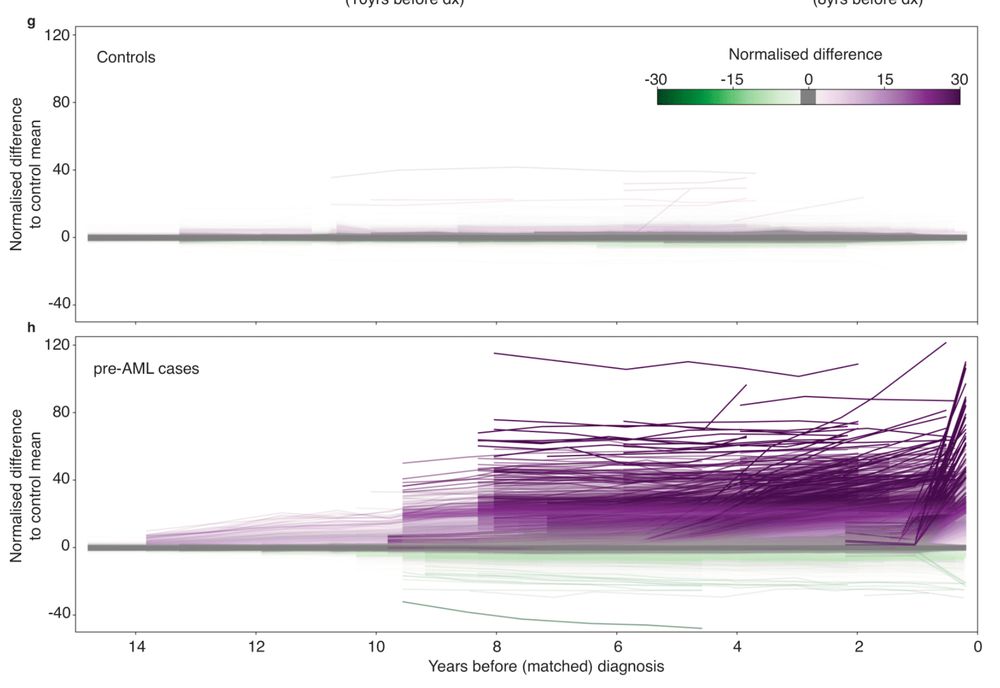
👉 Early cancer detection
👉 Methylation drivers
👉 Epimutation rates
👉 CpG lineage tracing
www.biorxiv.org/content/10.1...

shorturl.at/2LHbw
shorturl.at/2LHbw
www.science.org/doi/full/10....
www.science.org/doi/full/10....
Does the germline genome affect that predictability...?
Preprint: www.biorxiv.org/content/10.1...
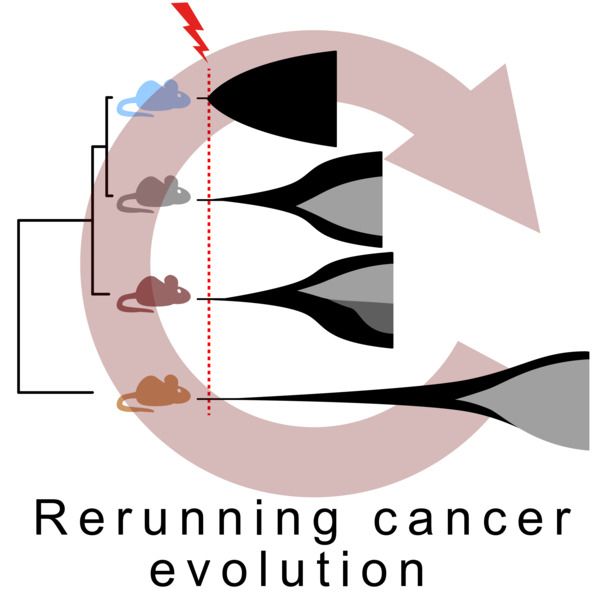
Does the germline genome affect that predictability...?
Preprint: www.biorxiv.org/content/10.1...