
Published in Genome Research @genomeresearch.bsky.social
Published in Genome Research @genomeresearch.bsky.social



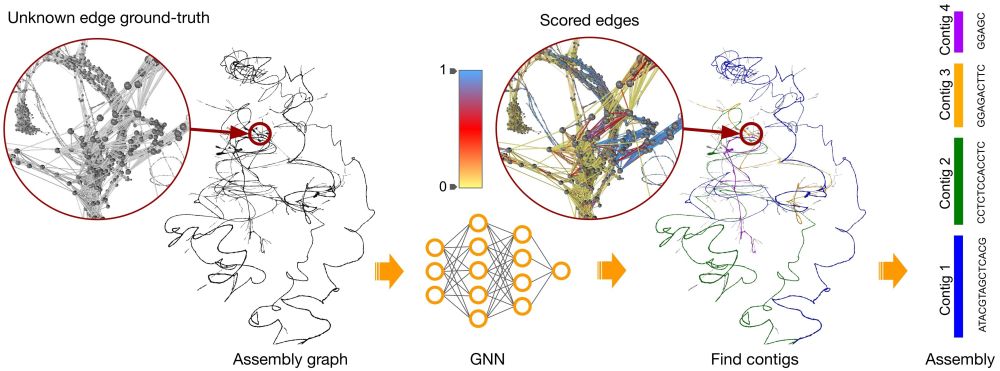

💻🧬 Scientists at Earlham Institute introduce MARTi, a new tool for real-time #nanopore #metagenomics.
@genomeresearch.bsky.social

💻🧬 Scientists at Earlham Institute introduce MARTi, a new tool for real-time #nanopore #metagenomics.
@genomeresearch.bsky.social
genome.cshlp.org/content/earl...
genome.cshlp.org/content/earl...




🧵 Check out my thread from yesterday for the details ⬇️
👨💻 Open source codes also available on my #GitHub page!
#AcademicSky
Our new study shows how to turn low-coverage data into valuable insights, analyzing >32,000 genomes from Belgium. #genetics #imputation #NIPS #AcademicSky
genome.cshlp.org/content/35/9...
🧵 Check out my thread from yesterday for the details ⬇️
👨💻 Open source codes also available on my #GitHub page!
#AcademicSky

www.su.se/department-o...

www.su.se/department-o...
Within the Solve_RD, @pacbio.bsky.social HiFi long-read whole-genome sequencing has proven instrumental in uncovering previously undetectable genetic variants, leading to new diagnoses
genome.cshlp.org/content/35/4...
@genomeresearch.bsky.social @erdera.bsky.social

Within the Solve_RD, @pacbio.bsky.social HiFi long-read whole-genome sequencing has proven instrumental in uncovering previously undetectable genetic variants, leading to new diagnoses
genome.cshlp.org/content/35/4...
@genomeresearch.bsky.social @erdera.bsky.social

About how #Wnt signalling activation induces #CTCF binding and formation of new #chromatin #loops at #CREs of target genes.
genome.cshlp.org/content/earl...
About how #Wnt signalling activation induces #CTCF binding and formation of new #chromatin #loops at #CREs of target genes.
genome.cshlp.org/content/earl...
📝 Now on @genomeresearch.bsky.social 👇
genome.cshlp.org/content/earl...
📝 Now on @genomeresearch.bsky.social 👇
genome.cshlp.org/content/earl...

