
MirGeneDB, MirMachine, MirMiner (soon!)





A presentation by the Argonautes Syndrom family alliance reminded how relevant research on Argonautes and related mechanisms is

A presentation by the Argonautes Syndrom family alliance reminded how relevant research on Argonautes and related mechanisms is
What to say: RNA U come around
What to say: RNA U come around



@arcecogen.bsky.social

@arcecogen.bsky.social
Parasitic flatworms also show massive microRNA losses (up to ~50%)
→ Suggesting a general principle in lophotrochozoan parasite evolution:
💥 Regulatory gene loss accompanies simplification.
pubmed.ncbi.nlm.nih.gov/24025793/
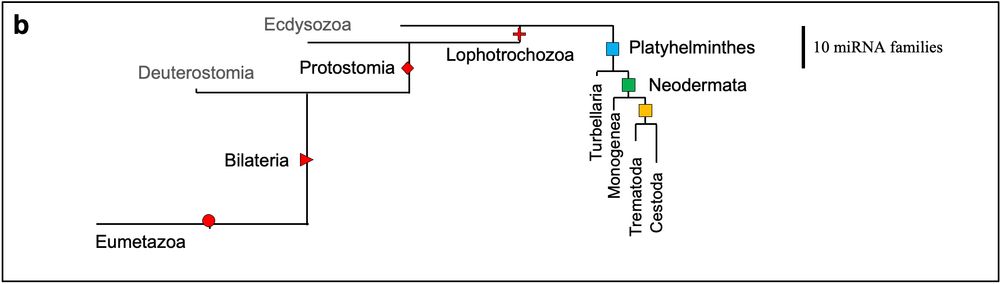
Parasitic flatworms also show massive microRNA losses (up to ~50%)
→ Suggesting a general principle in lophotrochozoan parasite evolution:
💥 Regulatory gene loss accompanies simplification.
pubmed.ncbi.nlm.nih.gov/24025793/
Losses:
▪️Digestive tract
▪️Ciliated head structures
▪️Protonephridia
And just like gene losses, morphological losses track with microRNA and BUSCO reductions.
Parasitism = evolutionary pruning. 🪓
However, new features evolve, too!
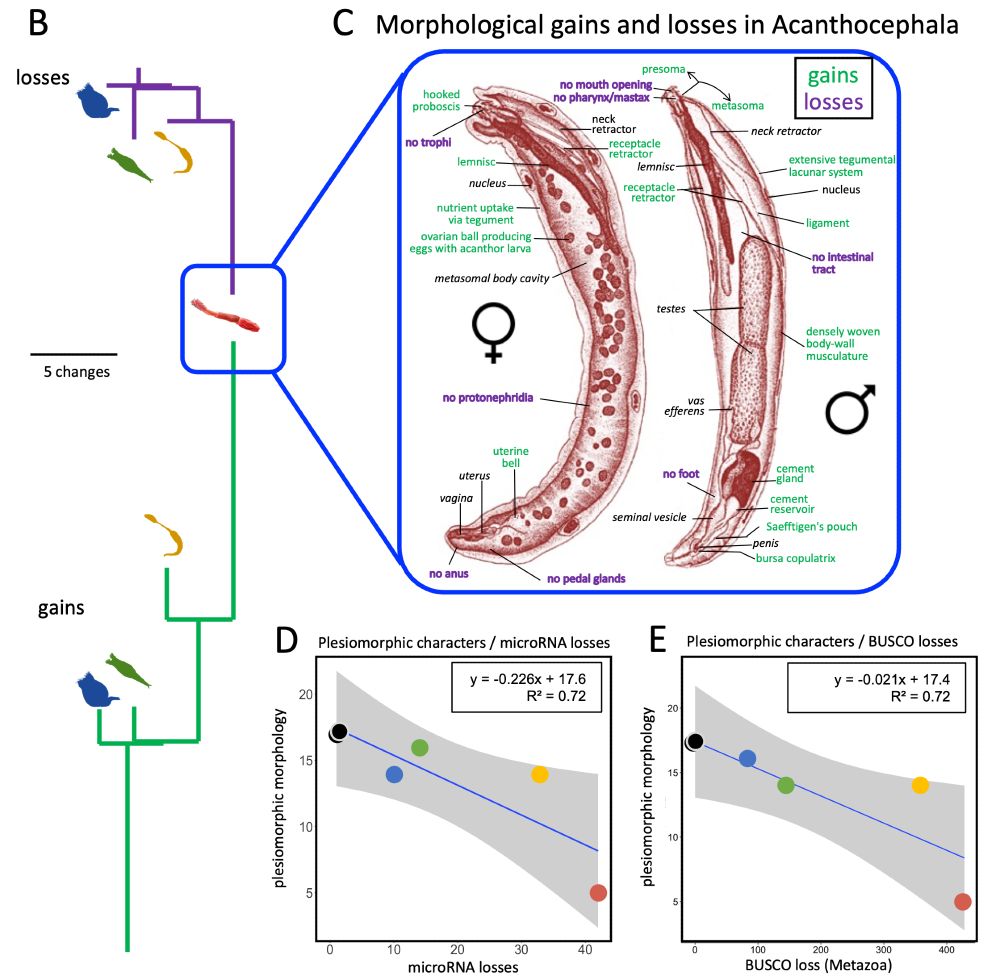
Losses:
▪️Digestive tract
▪️Ciliated head structures
▪️Protonephridia
And just like gene losses, morphological losses track with microRNA and BUSCO reductions.
Parasitism = evolutionary pruning. 🪓
However, new features evolve, too!
They're enriched for regulatory functions:
⚙️ Transcription factors
⚙️ RNA Pol II complex components
⚙️ Translation regulators
→ Suggests regulatory collapse as parasitism advances.
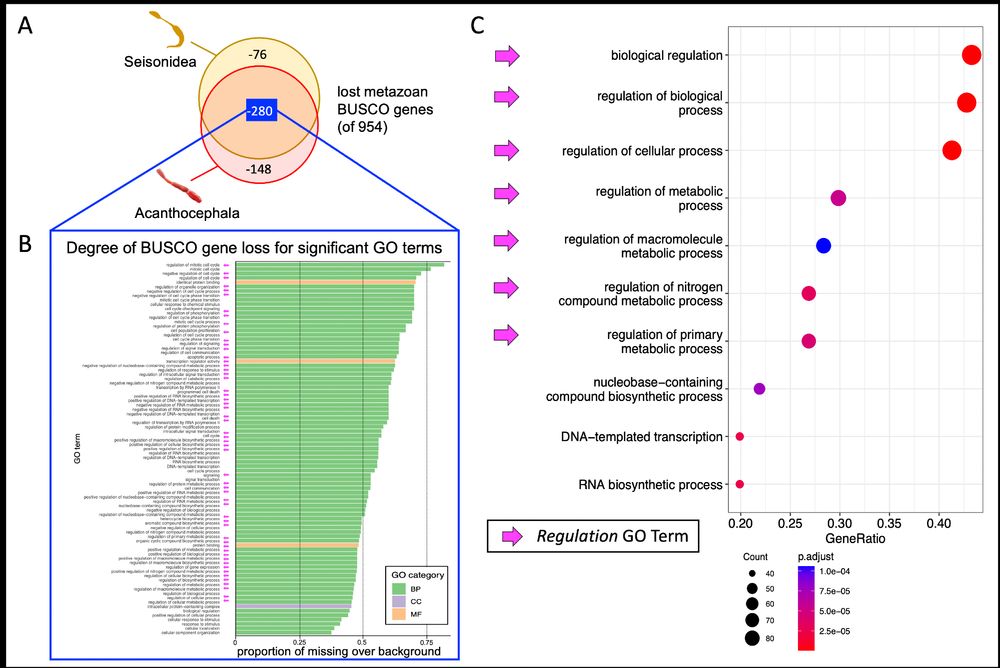
They're enriched for regulatory functions:
⚙️ Transcription factors
⚙️ RNA Pol II complex components
⚙️ Translation regulators
→ Suggests regulatory collapse as parasitism advances.
📉 Curiously we find that microRNA loss correlates with other forms of genome simplification.
🔸 Each lost microRNA family ≈ 400 protein-coding genes lost
🔸 Strong correlation with metazoan core gene loss (BUSCO)
🔸 Not explained by genome assembly quality or size
Simplification at every level.
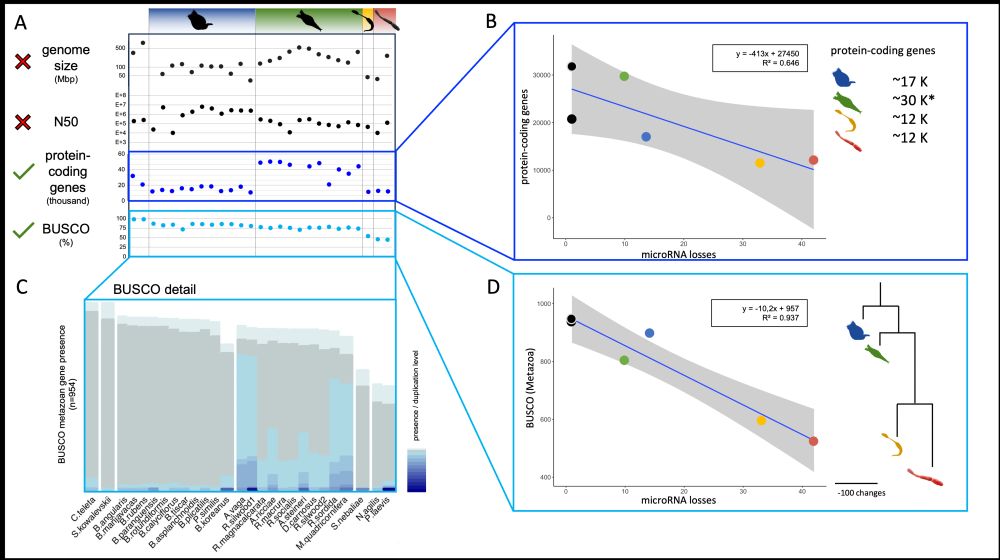
📉 Curiously we find that microRNA loss correlates with other forms of genome simplification.
🔸 Each lost microRNA family ≈ 400 protein-coding genes lost
🔸 Strong correlation with metazoan core gene loss (BUSCO)
🔸 Not explained by genome assembly quality or size
Simplification at every level.
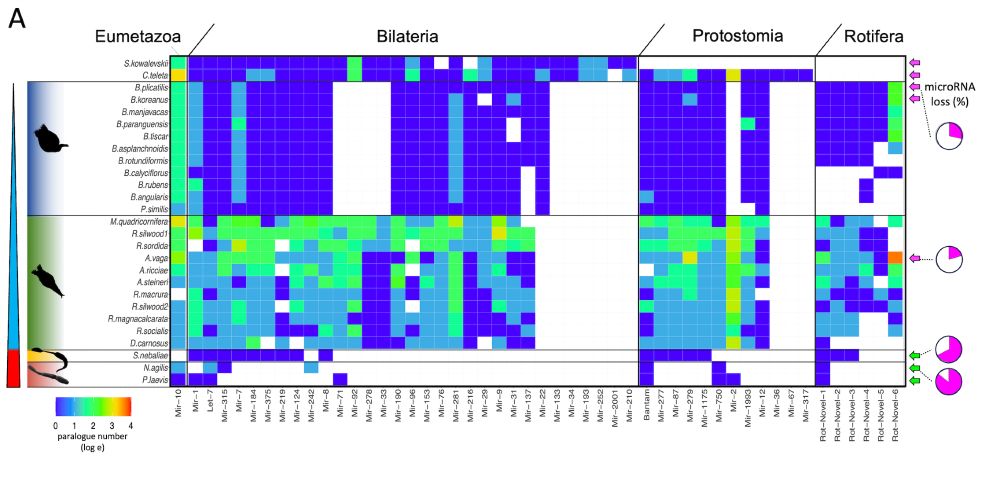
🧬 Figure 1: microRNA losses across Syndermata
🔹Free-living rotifers (Monogononta, Bdelloidea): mild miRNA loss
🔹Seisonidea: ~67% loss
🔹Acanthocephala: a shocking 85% microRNA family loss
This is the most extreme microRNA reduction ever reported in bilaterians.
📉🧫
🧬 Figure 1: microRNA losses across Syndermata
🔹Free-living rotifers (Monogononta, Bdelloidea): mild miRNA loss
🔹Seisonidea: ~67% loss
🔹Acanthocephala: a shocking 85% microRNA family loss
This is the most extreme microRNA reduction ever reported in bilaterians.
📉🧫

















