
Using programming & phylogenetics, I study & track viruses. Co-developer of Nextstrain, based at Swiss TPH & Uni Basel.
Founder of the orig SC2 variant-tracking website, CoVariants.org
I like 🐈 & 🛫
@[email protected]
Feedback? Questions? Drop a comment, DM or on GitHub. Like to join us in creating more EV datasets? Please reach out!
Thank you to everyone working on #Enteroviruses!🙏🏼
Feedback? Questions? Drop a comment, DM or on GitHub. Like to join us in creating more EV datasets? Please reach out!
Thank you to everyone working on #Enteroviruses!🙏🏼
This dataset was created by Nadia Neuner-Jehle, Alejandra Gonzalez Sanchez, & Emma Hodcroft - with help from Nextclade creators Richard Neher @neher.io & Ivan Aksamentov - and the input and advice of the ENPEN community!
This dataset was created by Nadia Neuner-Jehle, Alejandra Gonzalez Sanchez, & Emma Hodcroft - with help from Nextclade creators Richard Neher @neher.io & Ivan Aksamentov - and the input and advice of the ENPEN community!
We’re planning expansions - other non‐polio enteroviruses such as EV-A71, CVA10, CVA16.
🔭 Future features we hope to add: a recombination QC flag, gene-specific clade assignments.
Stay tuned & we welcome your input!
We’re planning expansions - other non‐polio enteroviruses such as EV-A71, CVA10, CVA16.
🔭 Future features we hope to add: a recombination QC flag, gene-specific clade assignments.
Stay tuned & we welcome your input!
You can also use the new EV-D68 dataset locally via the Nextclade CLI - perfect for integrating into workflows and sequencing pipelines!
Find out more about the CLI here: docs.nextstrain.org/projects/nex...
You can also use the new EV-D68 dataset locally via the Nextclade CLI - perfect for integrating into workflows and sequencing pipelines!
Find out more about the CLI here: docs.nextstrain.org/projects/nex...
- In the results: you’ll see QC status (red/yellow/white), clade calls, mutation counts, coverage gaps, etc. 🔠🚩💯
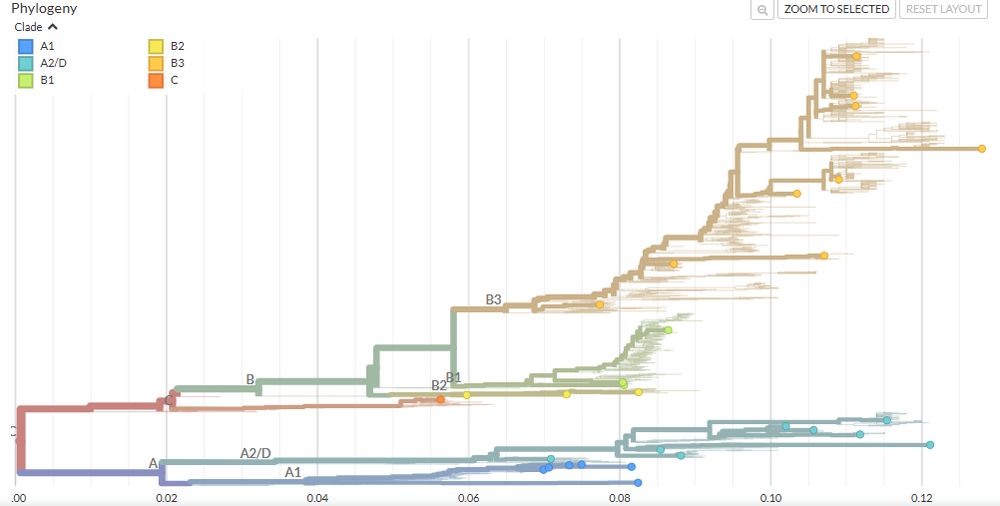
- The tree view places your sequences into the global phylogeny alongside curated reference data. 🌳

- In the results: you’ll see QC status (red/yellow/white), clade calls, mutation counts, coverage gaps, etc. 🔠🚩💯
- The tree view places your sequences into the global phylogeny alongside curated reference data. 🌳
- Go to nextclade.org → choose the EV-D68 dataset or go to clades.nextstrain.org?dataset-name...
- Drag & drop your FASTA (or load example)
- Click Run - alignment, QC & clade assignment happen in your browser (no data leaves your computer)

- Go to nextclade.org → choose the EV-D68 dataset or go to clades.nextstrain.org?dataset-name...
- Drag & drop your FASTA (or load example)
- Click Run - alignment, QC & clade assignment happen in your browser (no data leaves your computer)
- Clade assignments (A1–A2/D, B1–B3, C) 🔠
- Whole-genome mutation calling (not just VP1) Ⓜ️
- Quality-control (QC) metrics 🚩
- Gene annotations across the genome 🧬
👉🏻 So even if VP1 is missing, you can still assign the clade reliably.
- Clade assignments (A1–A2/D, B1–B3, C) 🔠
- Whole-genome mutation calling (not just VP1) Ⓜ️
- Quality-control (QC) metrics 🚩
- Gene annotations across the genome 🧬
👉🏻 So even if VP1 is missing, you can still assign the clade reliably.
📈 EV-D68 cases are already rising this season.
📂 This dataset lets clinicians & researchers quickly run clade assignments, mutation calls, and QC checks directly in their browser - with no data uploaded
Detailed info here!: eve-lab.org/blog/EV-D68-...

📈 EV-D68 cases are already rising this season.
📂 This dataset lets clinicians & researchers quickly run clade assignments, mutation calls, and QC checks directly in their browser - with no data uploaded
Detailed info here!: eve-lab.org/blog/EV-D68-...
But as a limited-contract, early-career scientist who spent a lot of pandemic night awake, terrified of being targeted, I can vouch that it's a tough & shit situation.
But as a limited-contract, early-career scientist who spent a lot of pandemic night awake, terrified of being targeted, I can vouch that it's a tough & shit situation.
And, there's interlinking at high levels. If there's friendships at govt level above you - you may not have any choice in what you can say publicly.
And, there's interlinking at high levels. If there's friendships at govt level above you - you may not have any choice in what you can say publicly.
Yes, the data is still available via GISAID website - but this is prohibited by DAA from display.
So GISAID decides what tools the public get.
Yes, the data is still available via GISAID website - but this is prohibited by DAA from display.
So GISAID decides what tools the public get.


