Franz Baumdicker
@fbaumdicker.bsky.social
Mathematical and Computational Population Genetics
Microbial Evolution | Machine Learning in PopGen | CRISPR-Cas | Pangenomes | HGT
🦠 ➔ 🧬 ➔ 💻 ➔ 🤔
www.baumdickergroup.de
Microbial Evolution | Machine Learning in PopGen | CRISPR-Cas | Pangenomes | HGT
🦠 ➔ 🧬 ➔ 💻 ➔ 🤔
www.baumdickergroup.de
Only a few hours left to submit your abstract for a talk at the Machine Learning in Evolutionary Genomics conference in December in Aussois in the French alps!

September 22, 2025 at 2:46 PM
Only a few hours left to submit your abstract for a talk at the Machine Learning in Evolutionary Genomics conference in December in Aussois in the French alps!
If you are interested in microbial #pangenomes, join us at @eseb2025.bsky.social in Barcelona with @bjesseshapiro.bsky.social and @annadewar.bsky.social
Only two days left to submit your abstract!
eseb2025.com/call-for-abs...
Only two days left to submit your abstract!
eseb2025.com/call-for-abs...

April 23, 2025 at 7:05 AM
If you are interested in microbial #pangenomes, join us at @eseb2025.bsky.social in Barcelona with @bjesseshapiro.bsky.social and @annadewar.bsky.social
Only two days left to submit your abstract!
eseb2025.com/call-for-abs...
Only two days left to submit your abstract!
eseb2025.com/call-for-abs...
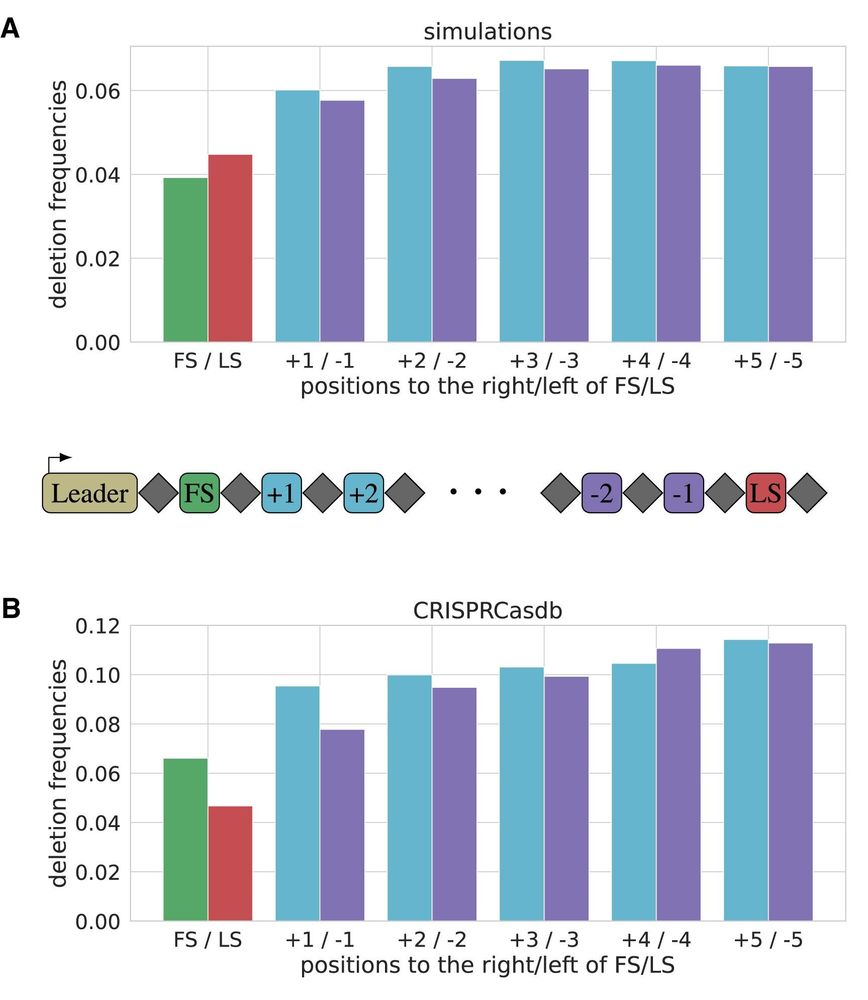
The spacer deletions at the very first and last positions of CRISPR arrays deviate from a symmetric boundary effect. The reconstructed deletion rate is higher at the first positions compared to the losses at the last positions. This might have multiple reasons ...
November 20, 2024 at 4:55 PM
The spacer deletions at the very first and last positions of CRISPR arrays deviate from a symmetric boundary effect. The reconstructed deletion rate is higher at the first positions compared to the losses at the last positions. This might have multiple reasons ...
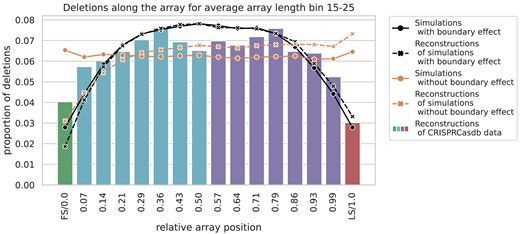
Consistent with this mechanism, we observe a “boundary effect”: Spacer deletions are less frequent near the leader and trailer ends of the array, potentially due to fewer neighboring repeats for alignment, while the middle of the array sees the most deletions.
November 20, 2024 at 4:55 PM
Consistent with this mechanism, we observe a “boundary effect”: Spacer deletions are less frequent near the leader and trailer ends of the array, potentially due to fewer neighboring repeats for alignment, while the middle of the array sees the most deletions.
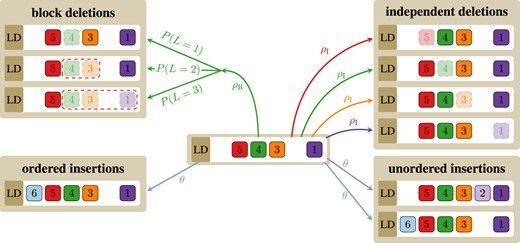
Interestingly, besides the loss of a single spacer, deletions occur often in blocks, involving an average of 2.7 spacers per event. This block deletion aligns with theories of replication slippage, where nearby repeats misalign, leading to multiple spacer deletions.
November 20, 2024 at 4:55 PM
Interestingly, besides the loss of a single spacer, deletions occur often in blocks, involving an average of 2.7 spacers per event. This block deletion aligns with theories of replication slippage, where nearby repeats misalign, leading to multiple spacer deletions.

