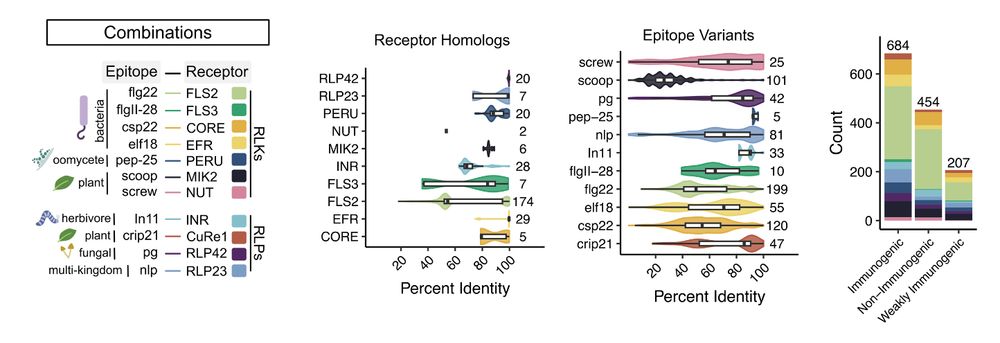
Previously immunogenicity was primarily characterized in the lab but was a expensive bottleneck considering the variation captured computationally!

Previously immunogenicity was primarily characterized in the lab but was a expensive bottleneck considering the variation captured computationally!