www.nature.com/articles/s41...

www.nature.com/articles/s41...

jobs.colorado.edu/jobs/JobDeta...
🧪🧬 #TESky #interferosky

jobs.colorado.edu/jobs/JobDeta...
🧪🧬 #TESky #interferosky
tinyurl.com/ydn6e3ac
tinyurl.com/ydn6e3ac
www.research.gov/grfp/Awardee...
www.research.gov/grfp/Awardee...
Gag proteins of endogenous retroviruses are required for zebrafish development
www.pnas.org/doi/10.1073/...
Led heroically by Sylvia Chang & @jonowells.bsky.social
A study which has changed the way I think of #transposons! No less! 🧵 1/n
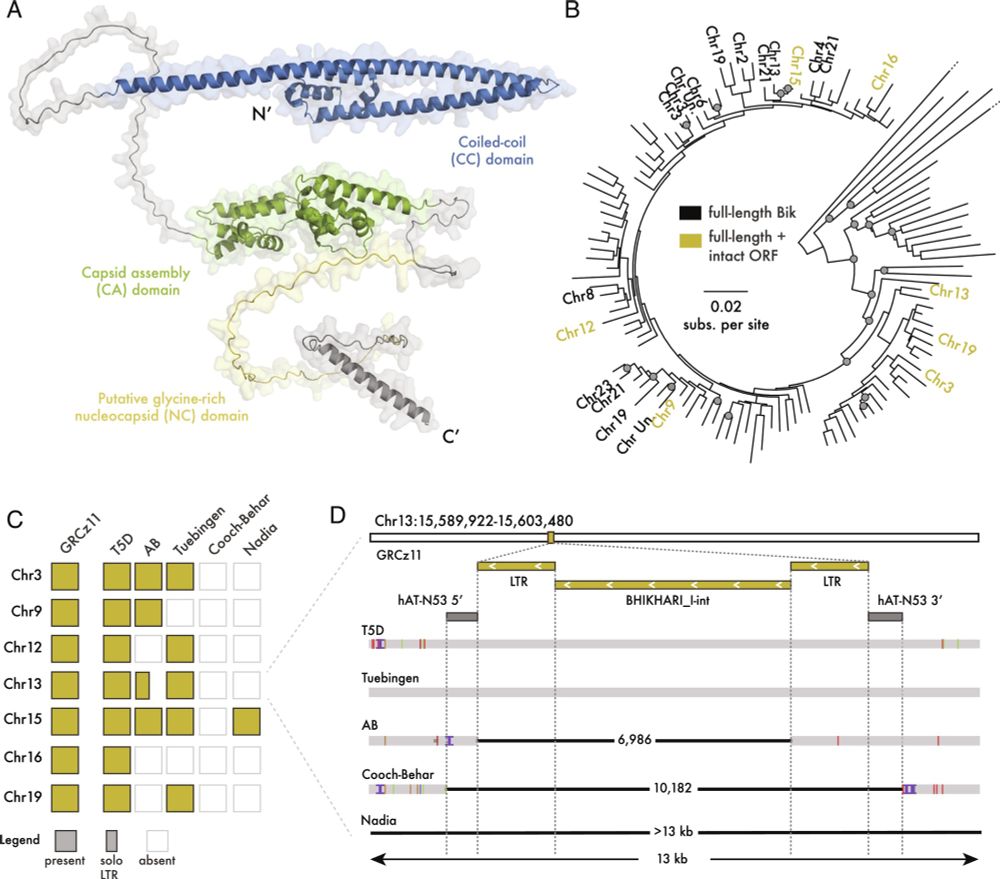
Gag proteins of endogenous retroviruses are required for zebrafish development
www.pnas.org/doi/10.1073/...
Led heroically by Sylvia Chang & @jonowells.bsky.social
A study which has changed the way I think of #transposons! No less! 🧵 1/n
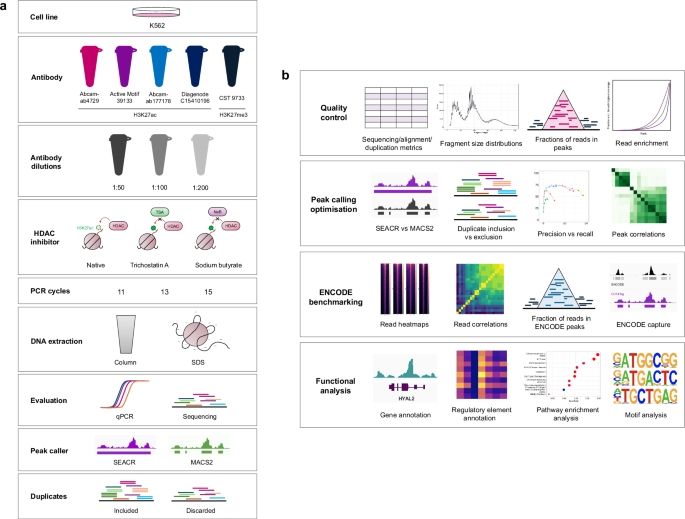
Excited to share our work on "Ab-trapping," an antibody artifact causing misleading peripheral ("rim") staining in imaging & genomics (IF, CUT&Tag, CUT&RUN). Antibodies fail to penetrate structures, accumulating at the periphery. A 🧵👇
doi.org/10.1101/2025...

Excited to share our work on "Ab-trapping," an antibody artifact causing misleading peripheral ("rim") staining in imaging & genomics (IF, CUT&Tag, CUT&RUN). Antibodies fail to penetrate structures, accumulating at the periphery. A 🧵👇
doi.org/10.1101/2025...
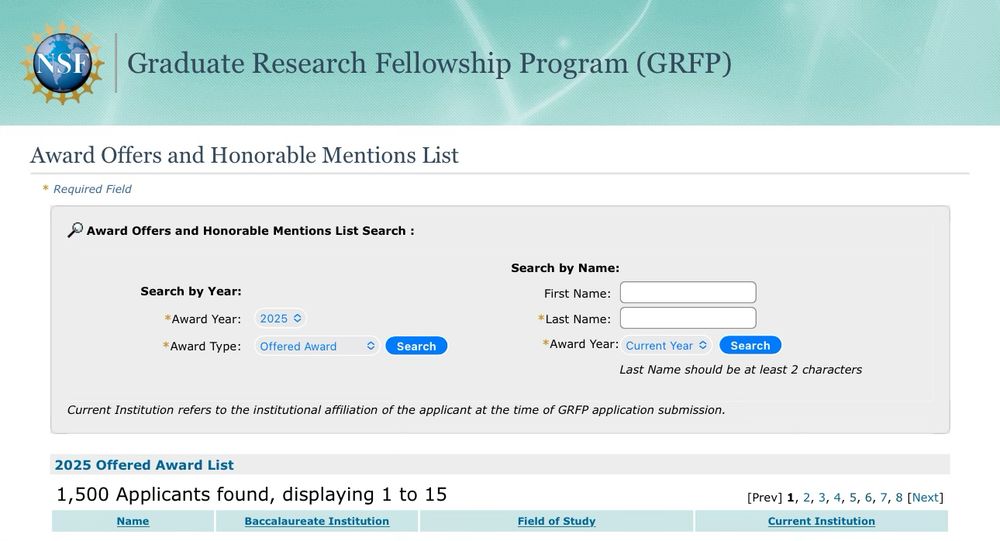
New graduate NSF fellowships will likely be announced this week for the GRFP (see bottom of 🧵 for why).
First off, I'm *very* glad to hear of those going forward, given the otherwise bleak year so far for graduate training.
Some observations... 1/12
New graduate NSF fellowships will likely be announced this week for the GRFP (see bottom of 🧵 for why).
First off, I'm *very* glad to hear of those going forward, given the otherwise bleak year so far for graduate training.
Some observations... 1/12
Loss of a single CTCF motif is sufficient to cause embryonic lethality.
www.sciencedirect.com/science/arti...
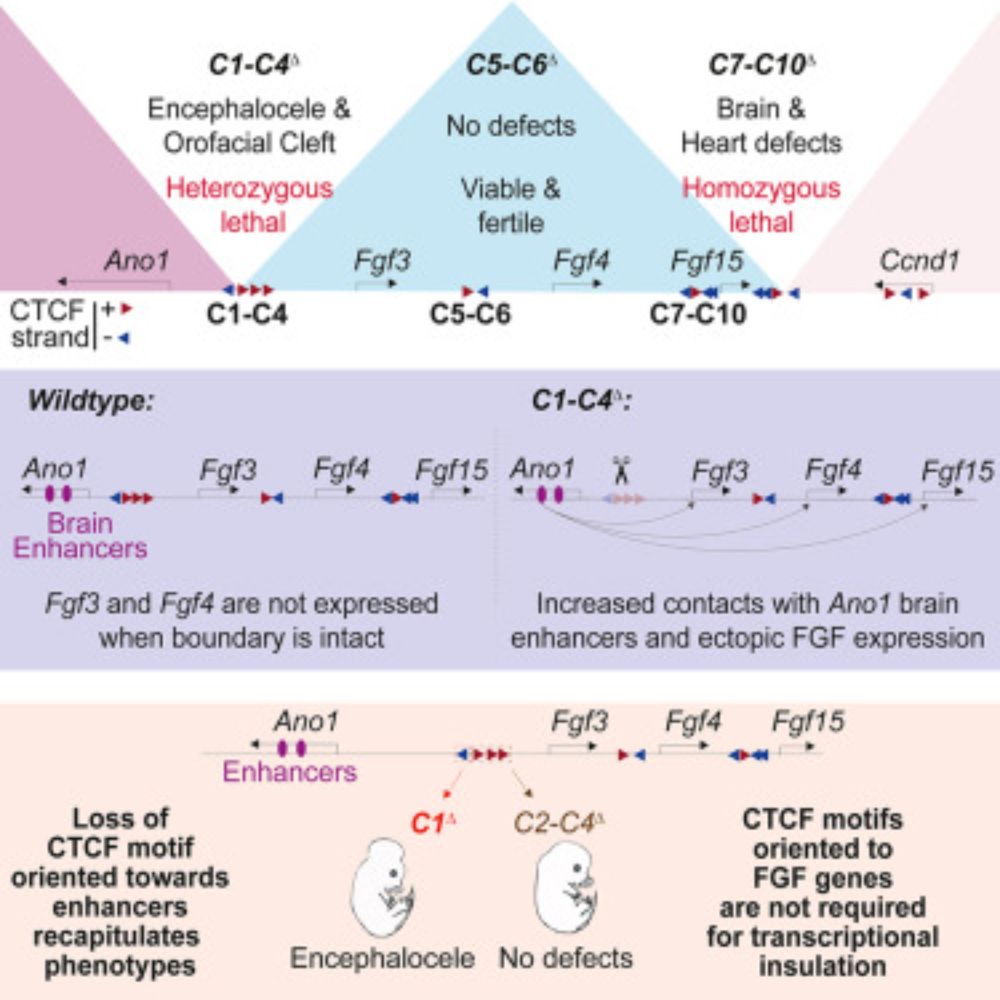
Loss of a single CTCF motif is sufficient to cause embryonic lethality.
www.sciencedirect.com/science/arti...
"Will the NSF Graduate Research Fellowship Program (GRFP) continue to award fellowships in 2025?"
GRFPs are a lifeline that support 2,000+ graduate students/year.
www.nsf.gov/executive-or...

"Will the NSF Graduate Research Fellowship Program (GRFP) continue to award fellowships in 2025?"
GRFPs are a lifeline that support 2,000+ graduate students/year.
www.nsf.gov/executive-or...
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
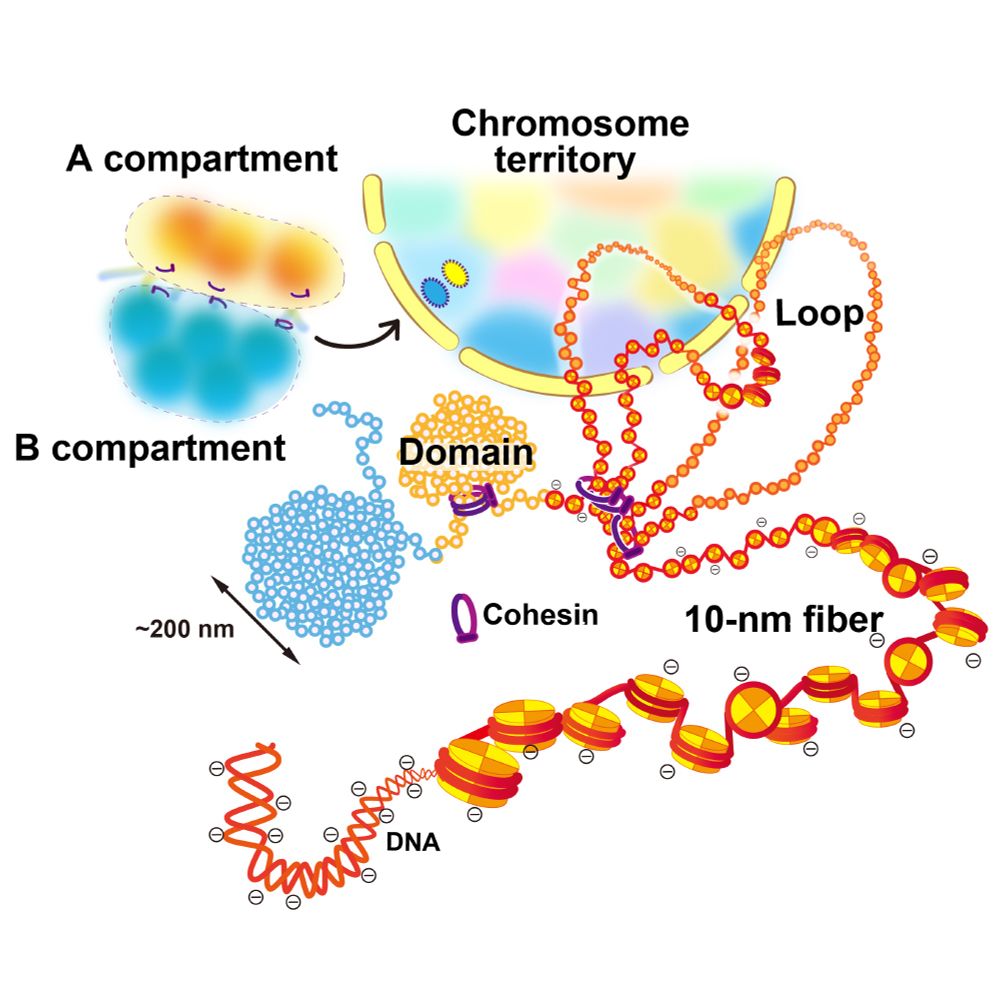
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
A short thread to highlight some of our findings 🧵
A short thread to highlight some of our findings 🧵
By integrating Micro-C with SuperRes Live-Imaging we can calibrate genomics&imaging to perform absolute quantification of looping (e.g. this loop is present 3%)
We quantify mESC 36k loops:
www.biorxiv.org/content/10.1...
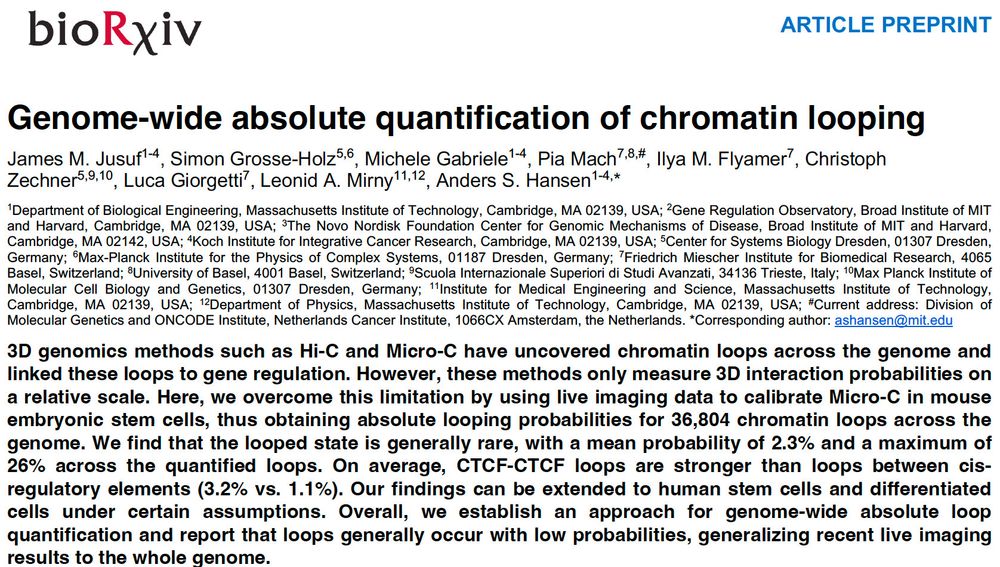

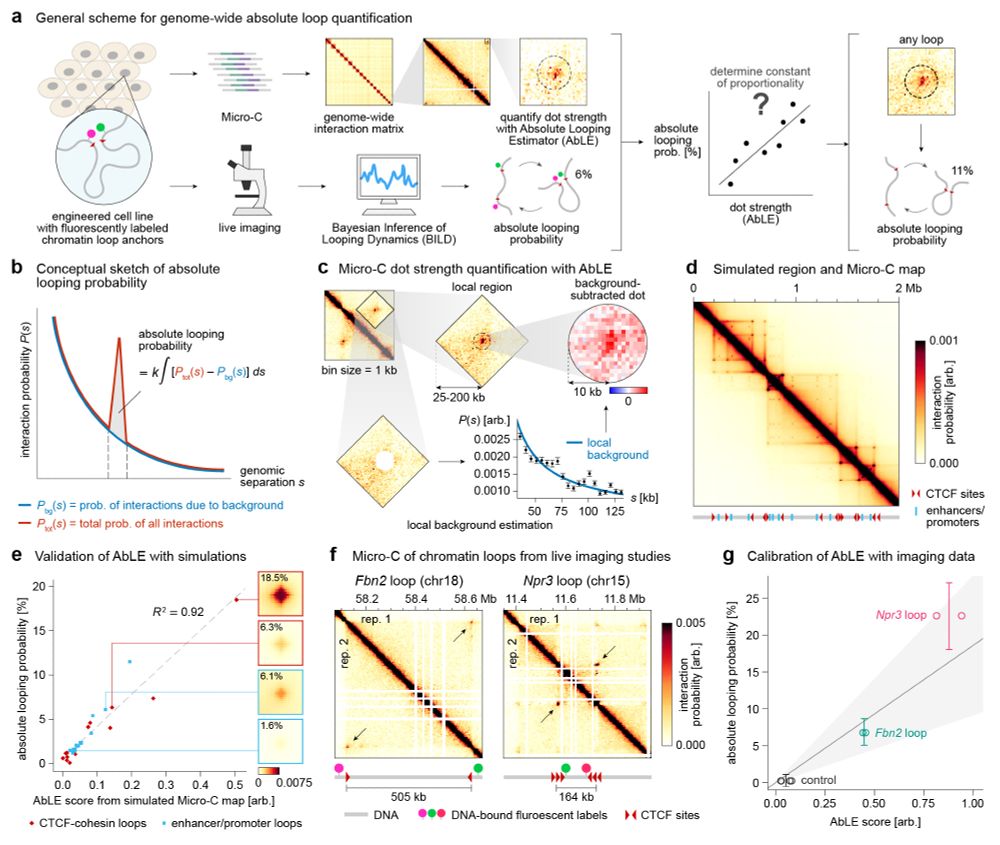
By integrating Micro-C with SuperRes Live-Imaging we can calibrate genomics&imaging to perform absolute quantification of looping (e.g. this loop is present 3%)
We quantify mESC 36k loops:
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Special thanks to @aliceyting.bsky.social for discussions
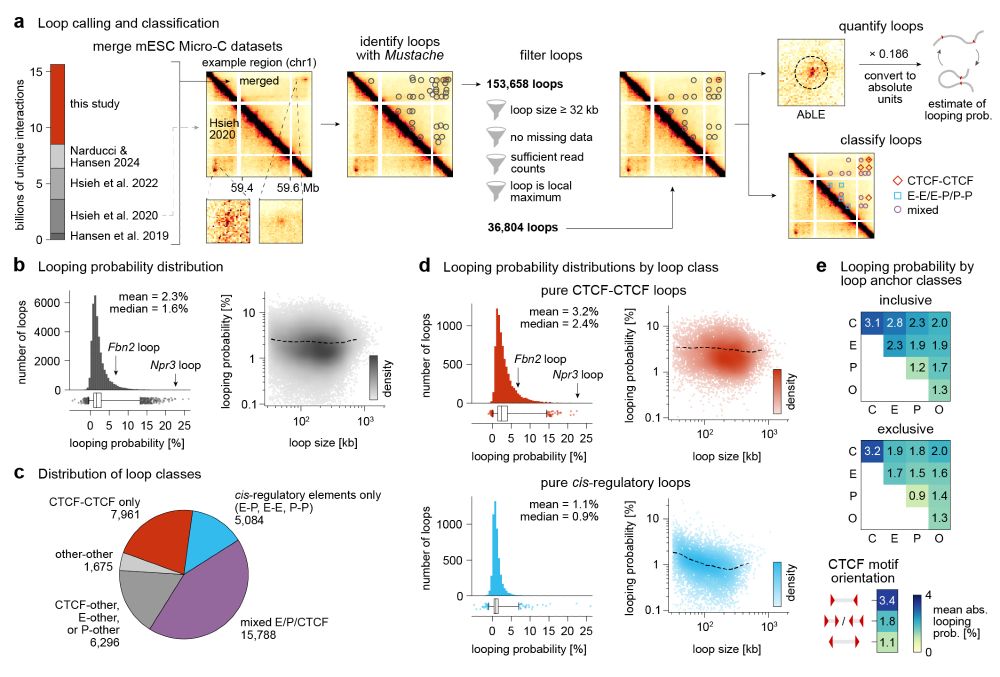
www.biorxiv.org/content/10.1...
Special thanks to @aliceyting.bsky.social for discussions
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...

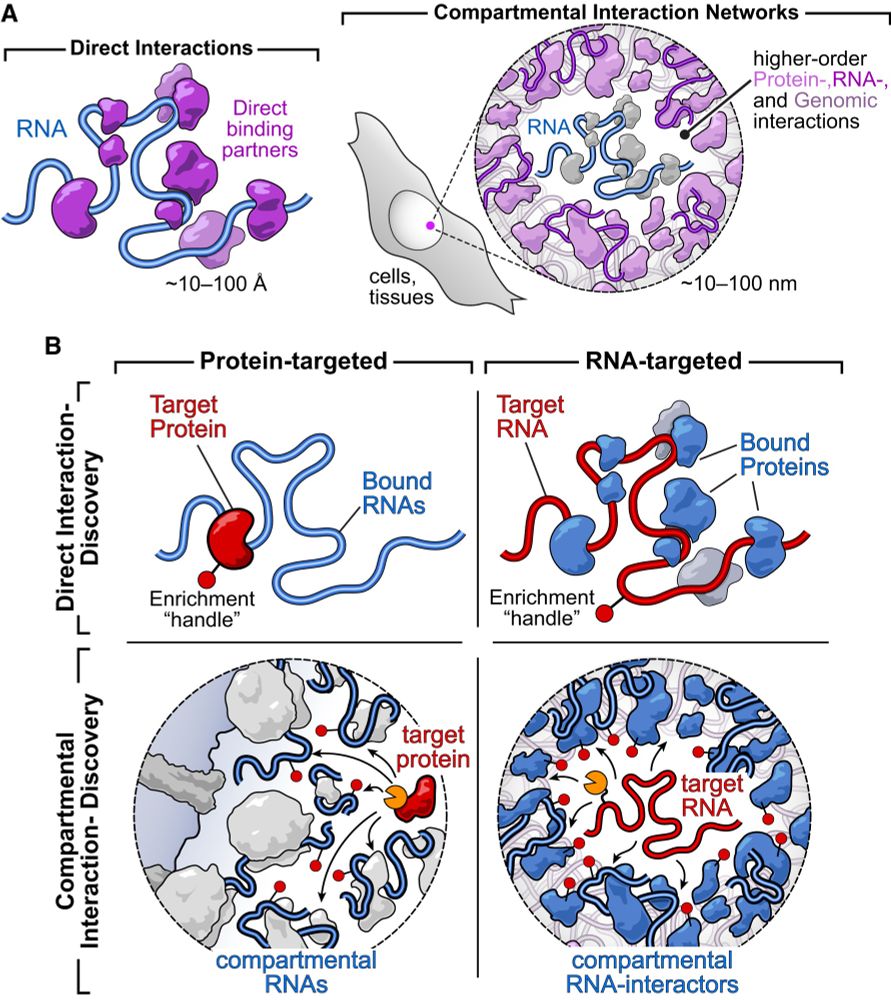
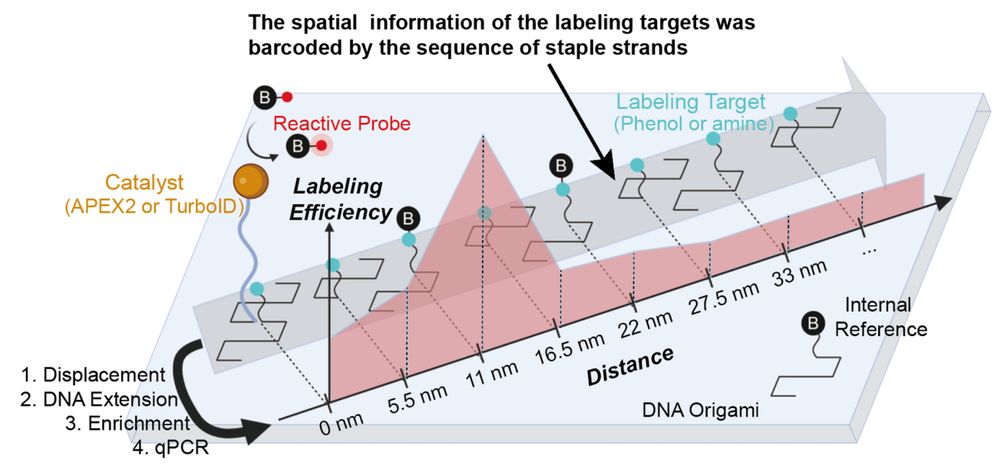
If you've ever wanted to dissect the subnuclear "neighborhood" around an individual locus, read on! (1/30)
If you've ever wanted to dissect the subnuclear "neighborhood" around an individual locus, read on! (1/30)
go.bsky.app/L8RAbiJ
It's fairly sparse for now, so please let me know if you'd like to be added!

go.bsky.app/L8RAbiJ
It's fairly sparse for now, so please let me know if you'd like to be added!
www.biorxiv.org/content/10.1...
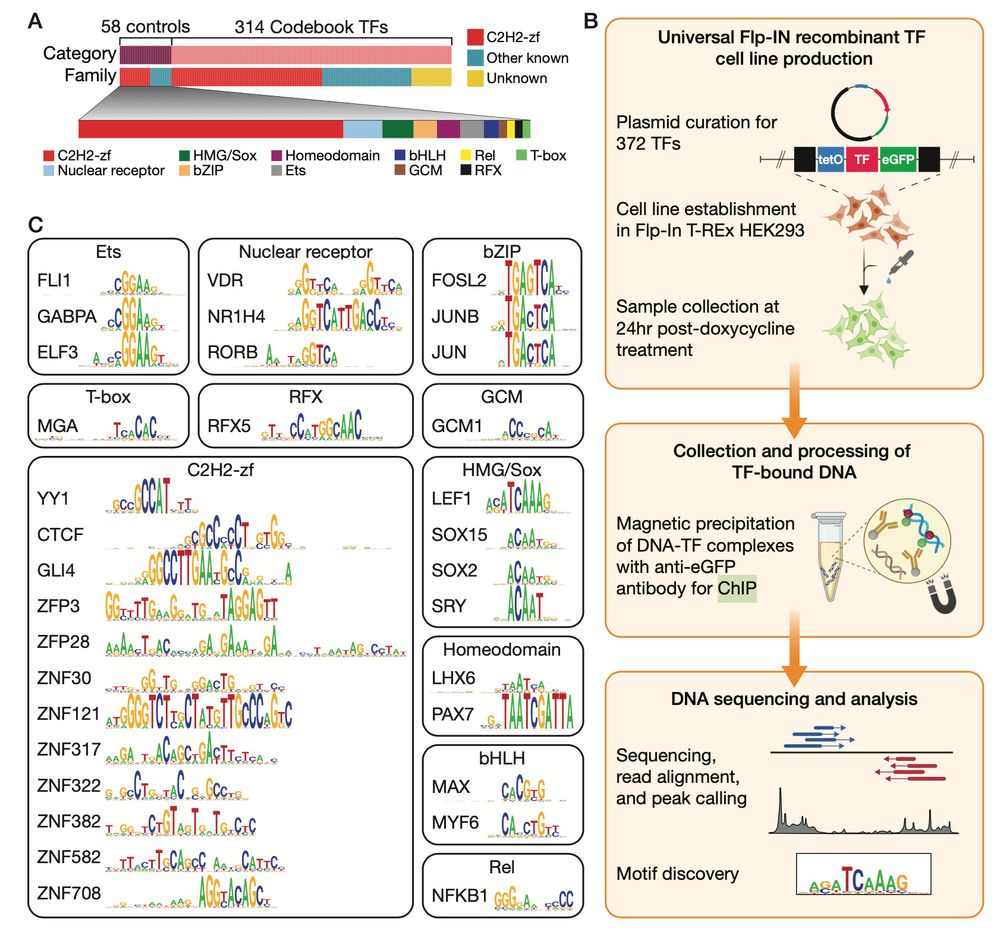
www.biorxiv.org/content/10.1...
pSABER is a new method for amplifying in situ hybridization signals
This work was driven by Sahar Attar and co-led by Ram Akliesh ([at] podocytes on the other app) with help from @shechnerlab.bsky.social @dschweppe.bsky.social
doi.org/10.1038/s415...
🧵 1/12

pSABER is a new method for amplifying in situ hybridization signals
This work was driven by Sahar Attar and co-led by Ram Akliesh ([at] podocytes on the other app) with help from @shechnerlab.bsky.social @dschweppe.bsky.social
doi.org/10.1038/s415...
🧵 1/12

