
Formerly at Sanger institute working on recessive developmental disorders in DDD
Probably with a sd of 20 😂
Paying £40k for that is insanity
Probably with a sd of 20 😂
Paying £40k for that is insanity
I’m still in team “2nd tier test” 😅 but all of your work has made me think about it more.
Do you think you will make much gain using pangenome assembly, in particular for complex gene/regions?
I’m still in team “2nd tier test” 😅 but all of your work has made me think about it more.
Do you think you will make much gain using pangenome assembly, in particular for complex gene/regions?
And for All of Us v8, we will provide you with the QC’ed pgen files on publication in a public workspace available to registered users.

And for All of Us v8, we will provide you with the QC’ed pgen files on publication in a public workspace available to registered users.
This time we didn’t ignore the X chromosome! We show that you should pay special attention to non-pseudoautosomal X chromosome where QC should be more lenient for haploid males.
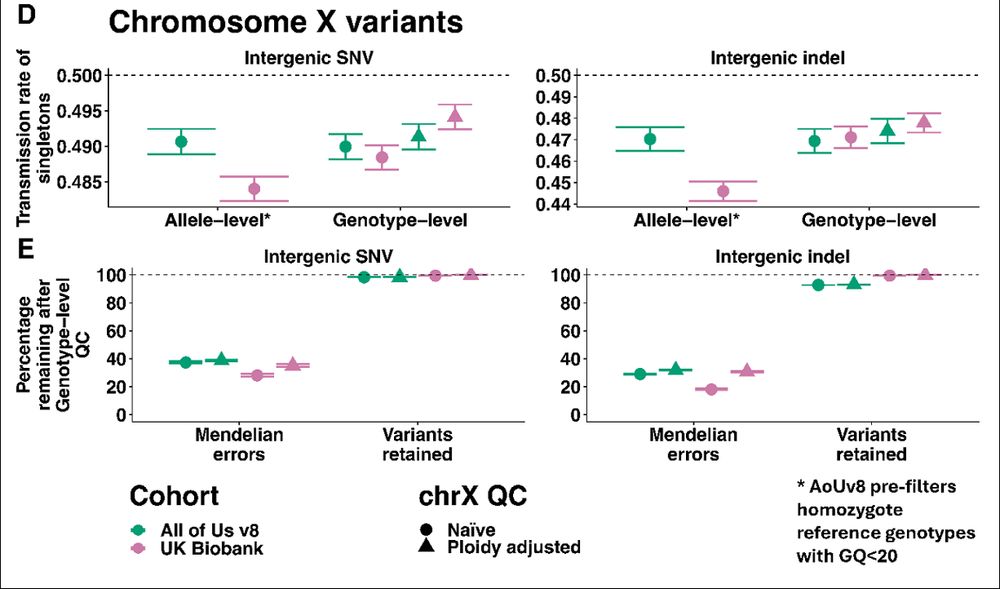
This time we didn’t ignore the X chromosome! We show that you should pay special attention to non-pseudoautosomal X chromosome where QC should be more lenient for haploid males.
Trios! Both cohorts have ~1k parent-offspring trios that were recruited incidentally.
Applying genotype-level QC reduces Mendelian errors by ~60-80% (even in All of Us where they already did genotype-level QC on hom-refs!)

Trios! Both cohorts have ~1k parent-offspring trios that were recruited incidentally.
Applying genotype-level QC reduces Mendelian errors by ~60-80% (even in All of Us where they already did genotype-level QC on hom-refs!)
After genotype-level QC and a 10% missingness cut-off, we remove ~100 million (~9%) variants!
Most genotypes removed are homozygote reference (which were filtered in All of Us already)

After genotype-level QC and a 10% missingness cut-off, we remove ~100 million (~9%) variants!
Most genotypes removed are homozygote reference (which were filtered in All of Us already)
Here, we show how we determine data quality in WGS data, provide a really fast way of doing so on biobank data, and we will release QC’ed plink files for All of Us upon publication

Here, we show how we determine data quality in WGS data, provide a really fast way of doing so on biobank data, and we will release QC’ed plink files for All of Us upon publication

