mysite.science.uottawa.ca/rchica/
See below for an early example:
www.pnas.org/doi/full/10....

See below for an early example:
www.pnas.org/doi/full/10....
www.nature.com/articles/nch...
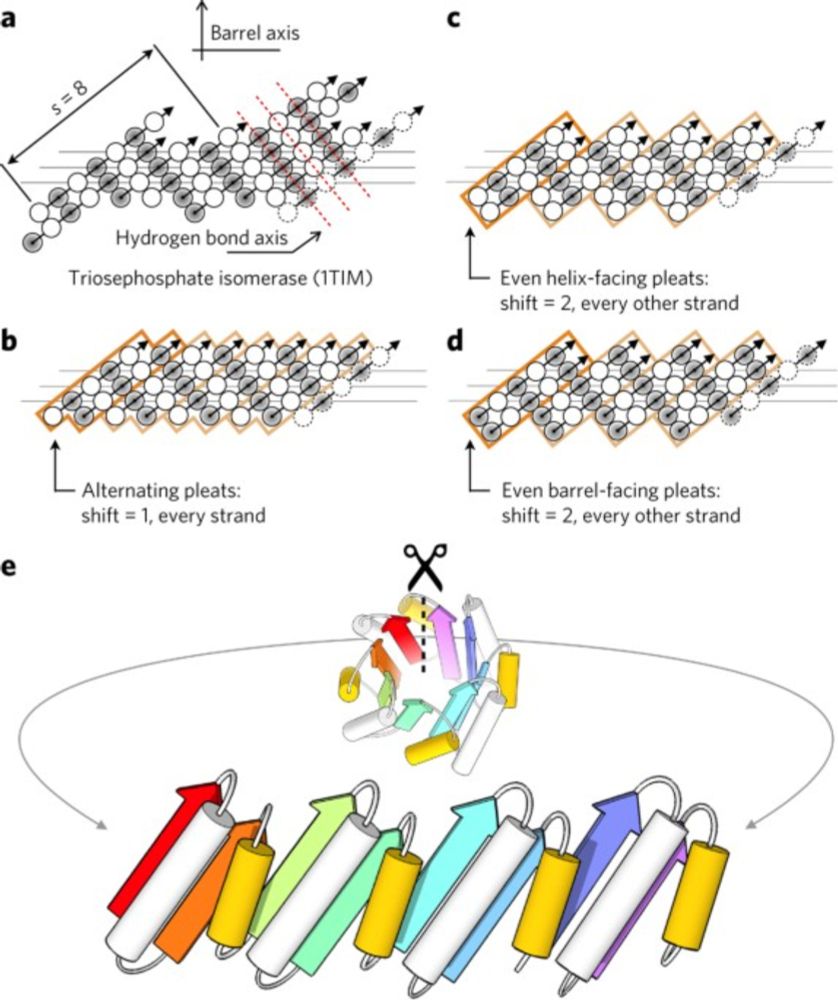
www.nature.com/articles/nch...
An early pioneer of this field is Bill DeGrado, see below.
www.science.org/doi/10.1126/...

An early pioneer of this field is Bill DeGrado, see below.
www.science.org/doi/10.1126/...
✅ Introduces a new strategy to transform minimal protein scaffolds into biocatalysts
✅ Provides mechanistic insights from crystallography & molecular dynamics
✅ Opens the door to designing custom lids for more complex reactions, which we’re now exploring
Thanks for reading! 🧵🧬
✅ Introduces a new strategy to transform minimal protein scaffolds into biocatalysts
✅ Provides mechanistic insights from crystallography & molecular dynamics
✅ Opens the door to designing custom lids for more complex reactions, which we’re now exploring
Thanks for reading! 🧵🧬
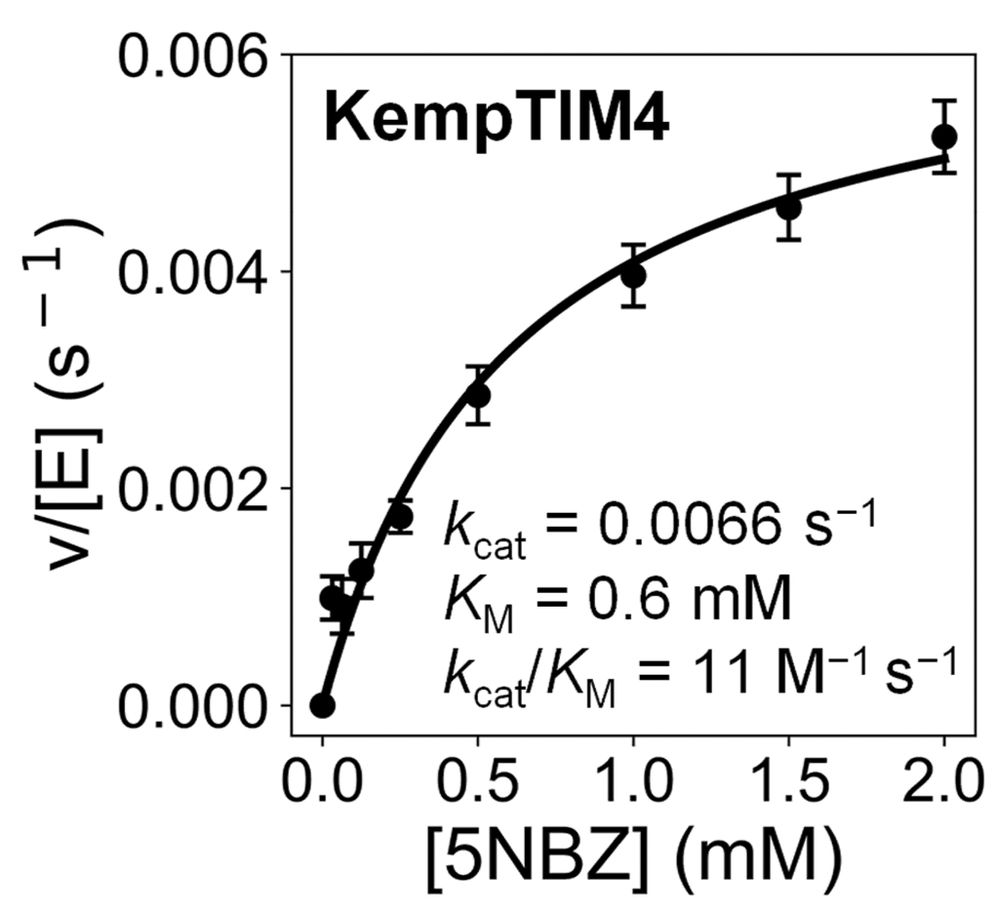
However, a subtle 1.8 Å lid shift disrupted a key catalytic contact, likely contributing to the modest activity. But structural analysis reveals paths to improve activity in the next round of design!
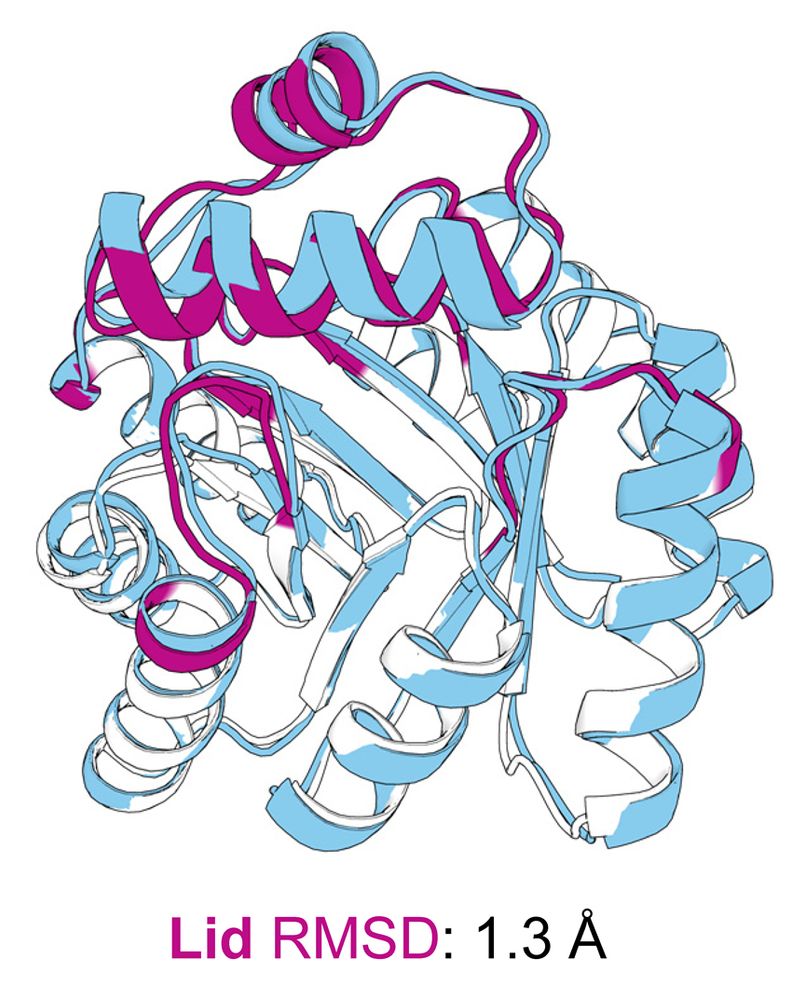
However, a subtle 1.8 Å lid shift disrupted a key catalytic contact, likely contributing to the modest activity. But structural analysis reveals paths to improve activity in the next round of design!
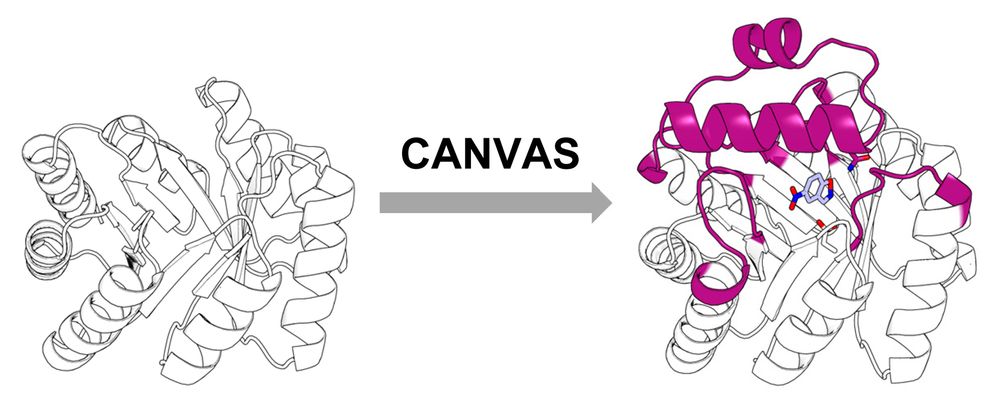
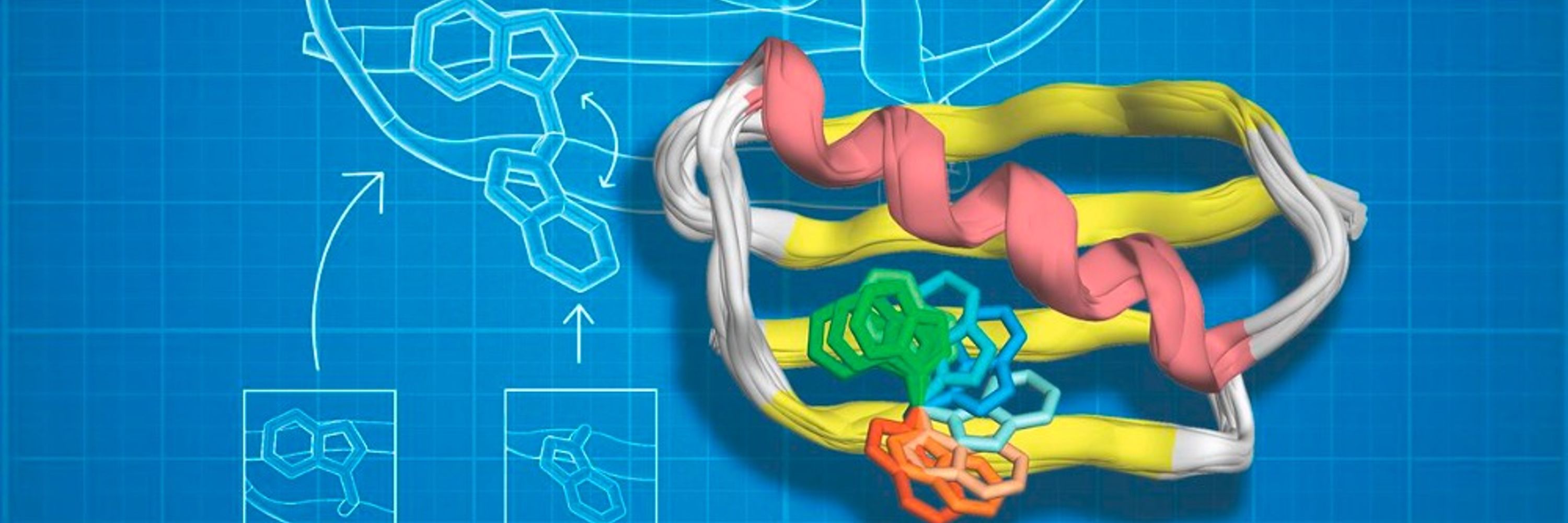
Enter CANVAS: a computational pipeline combining Triad, RFdiffusion & ProteinMPNN to customize minimal TIM barrels into functional enzymes.
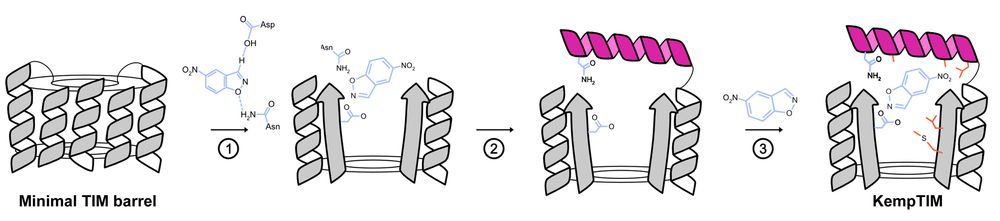
Enter CANVAS: a computational pipeline combining Triad, RFdiffusion & ProteinMPNN to customize minimal TIM barrels into functional enzymes.
(6/6)

(6/6)
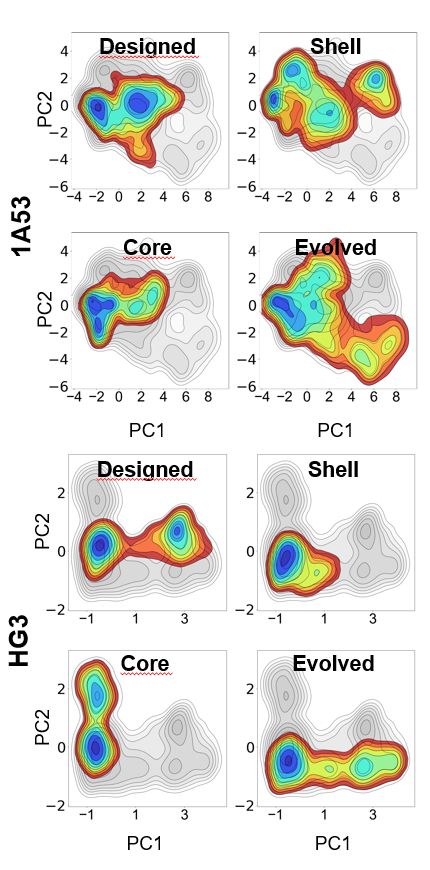

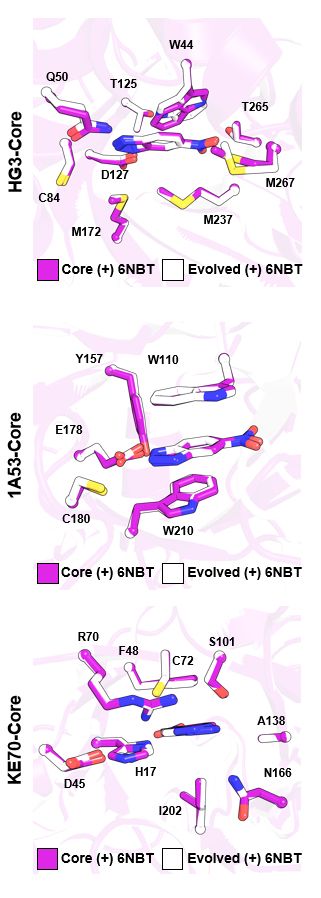
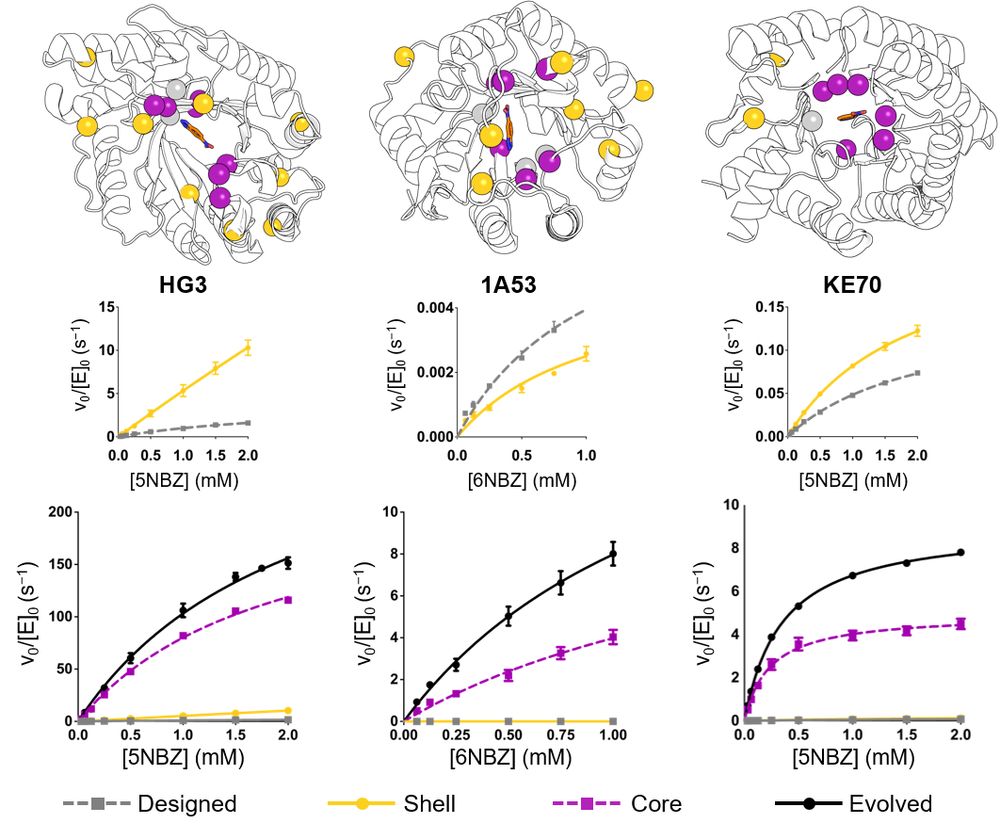
What does this mean for enzyme design? Find out in our preprint: www.biorxiv.org/content/10.1...
#Enzymology #Biophysics #Catalysis #EnzymeDesign

What does this mean for enzyme design? Find out in our preprint: www.biorxiv.org/content/10.1...
#Enzymology #Biophysics #Catalysis #EnzymeDesign

