#NewPI #DiversityEqualityInclusion
Single-cell, multi-omics, machine learning
www.nature.com/articles/s41...

www.nature.com/articles/s41...
STORIES learns cell fate landscapes from spatial tramscripromics data profiled at several time points, thus allowing prediction of future cell states.
Led by Geert-Jan Huizing and Jules Samaran
www.nature.com/articles/s41...
@pasteur.fr

STORIES learns cell fate landscapes from spatial tramscripromics data profiled at several time points, thus allowing prediction of future cell states.
Led by Geert-Jan Huizing and Jules Samaran
www.nature.com/articles/s41...
@pasteur.fr
It was really interesting to write a short manuscript mixing method presentation & broader insights on data preprocessing. 📝
Thank you to my co-supervisors @cantinilab.bsky.social & @juliosaezrod.bsky.social for their feedback on this side work! 🫶
5/5

Available here: github.com/gletort/Fish... user documentation: gletort.github.io/FishFeats/
Available here: github.com/gletort/Fish... user documentation: gletort.github.io/FishFeats/
To predict the behaviour of a primate, would you rather base your guess on a closely related species or one with a similar brain shape? We looked at brains & behaviours of 70 species, you’ll be surprised!
🧵Thread on our new preprint with @r3rt0.bsky.social , doi.org/10.1101/2025...
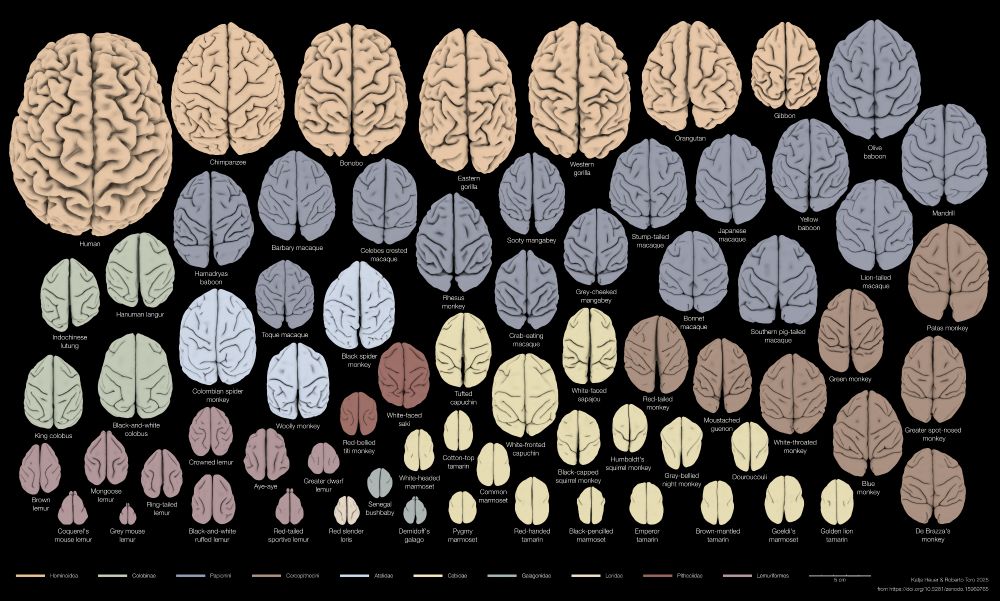
To predict the behaviour of a primate, would you rather base your guess on a closely related species or one with a similar brain shape? We looked at brains & behaviours of 70 species, you’ll be surprised!
🧵Thread on our new preprint with @r3rt0.bsky.social , doi.org/10.1101/2025...
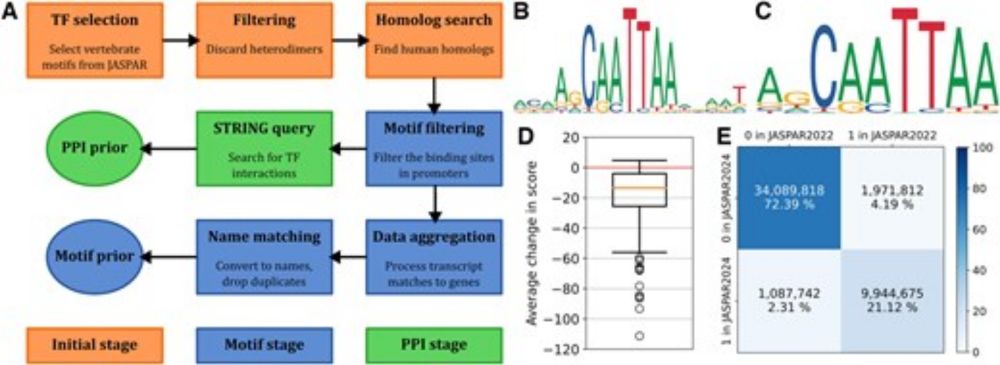
If you are interested in multi-omics matrix factorization when few samples are available (e.g. rare diseases), have a look at it!
#multiomics #machinelearning #transferlearning
🧬 When you only have a few datasets available for multi-omics integration, for instance because you work on rare diseases, one solution is Transfer Learning. Introducing #MOTL - now available in Genome Biology!
doi.org/10.1186/s130...
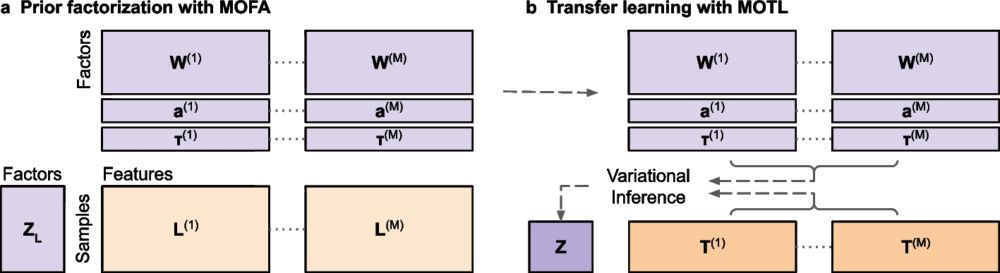
If you are interested in multi-omics matrix factorization when few samples are available (e.g. rare diseases), have a look at it!
#multiomics #machinelearning #transferlearning
Thanks again @anaisbaudot.bsky.social and all #NetBio commitee for the invitation!
@institutpasteur.bsky.social #singlecell #AI #netbio
She is sharing cutting-edge work on multi-modal learning for single-cell data integration, tackling how to combine diverse omics and spatial data to unlock new insights into cellular heterogeneity.

Thanks again @anaisbaudot.bsky.social and all #NetBio commitee for the invitation!
@institutpasteur.bsky.social #singlecell #AI #netbio
Interested and attending #ISMBECCB2025? Let’s connect!
🔗 www.jobbnorge.no/en/available...

Interested and attending #ISMBECCB2025? Let’s connect!
🔗 www.jobbnorge.no/en/available...
She is sharing cutting-edge work on multi-modal learning for single-cell data integration, tackling how to combine diverse omics and spatial data to unlock new insights into cellular heterogeneity.

She is sharing cutting-edge work on multi-modal learning for single-cell data integration, tackling how to combine diverse omics and spatial data to unlock new insights into cellular heterogeneity.
Join us at the @ncmbm.bsky.social, part of the @nordicembl.bsky.social, at @uio.no
Feel free to reach out if you have Qs.
🧬 With an attractive start-up package, help us shape the future of molecular biosciences and medicine in 🇳🇴 and be part of the @nordicembl.bsky.social!

Join us at the @ncmbm.bsky.social, part of the @nordicembl.bsky.social, at @uio.no
Feel free to reach out if you have Qs.
#ParisSaclaySPRING #SPRING25



#ParisSaclaySPRING #SPRING25
#NetBioMed2025 #NetSci2025 @verapancaldi.bsky.social



#NetBioMed2025 #NetSci2025 @verapancaldi.bsky.social

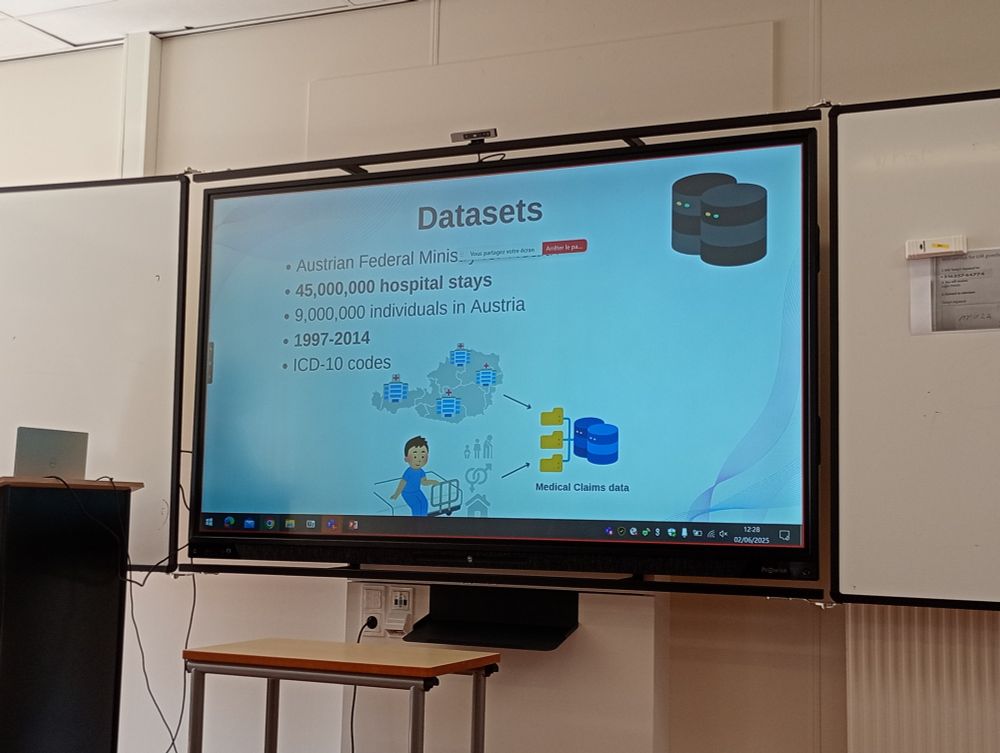
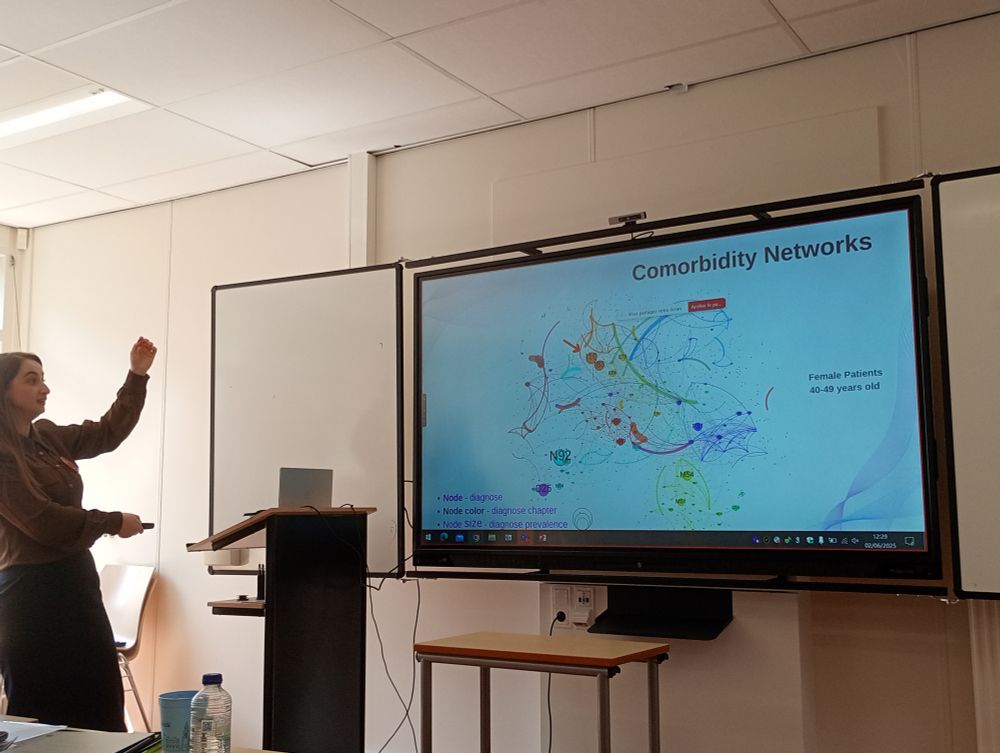


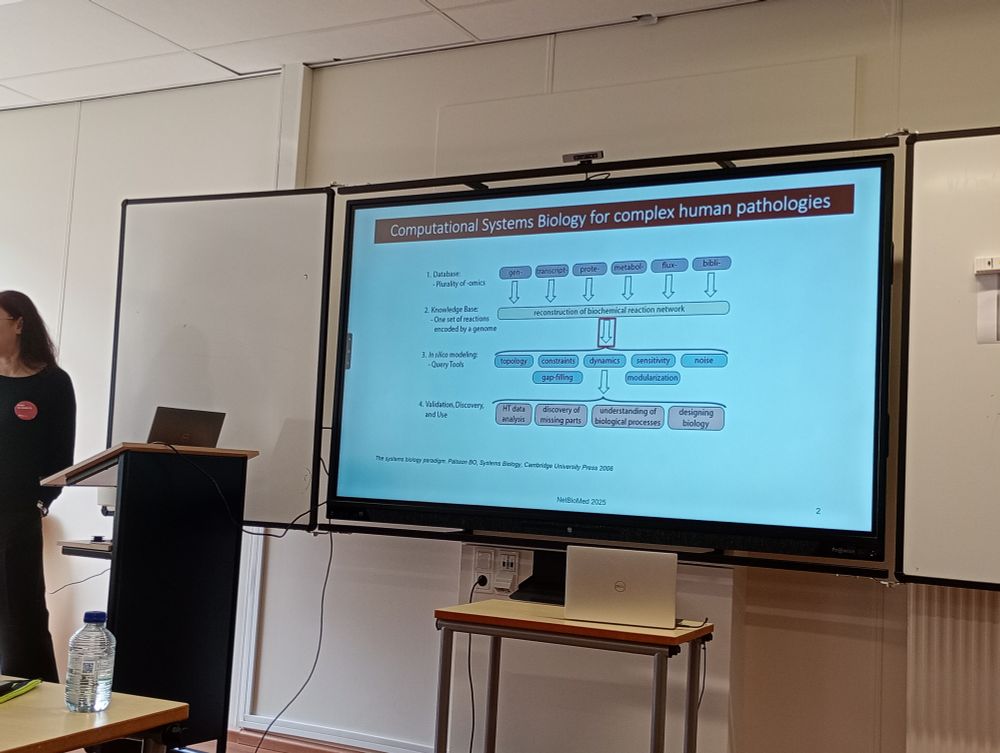

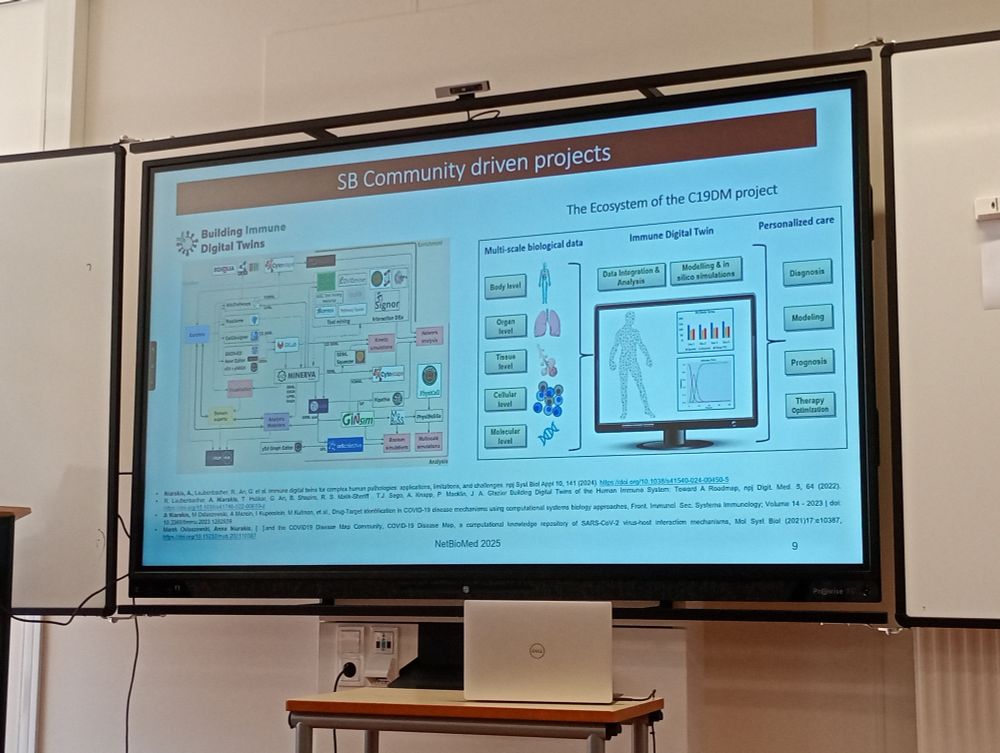
#NetBioMed2025 #NetSci2025

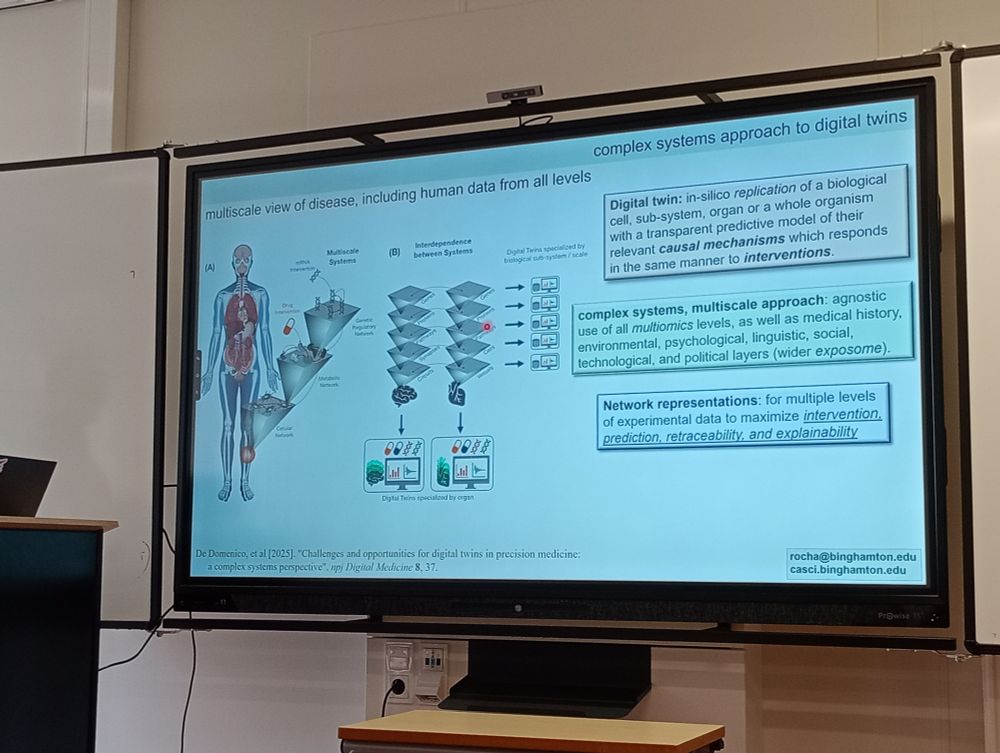
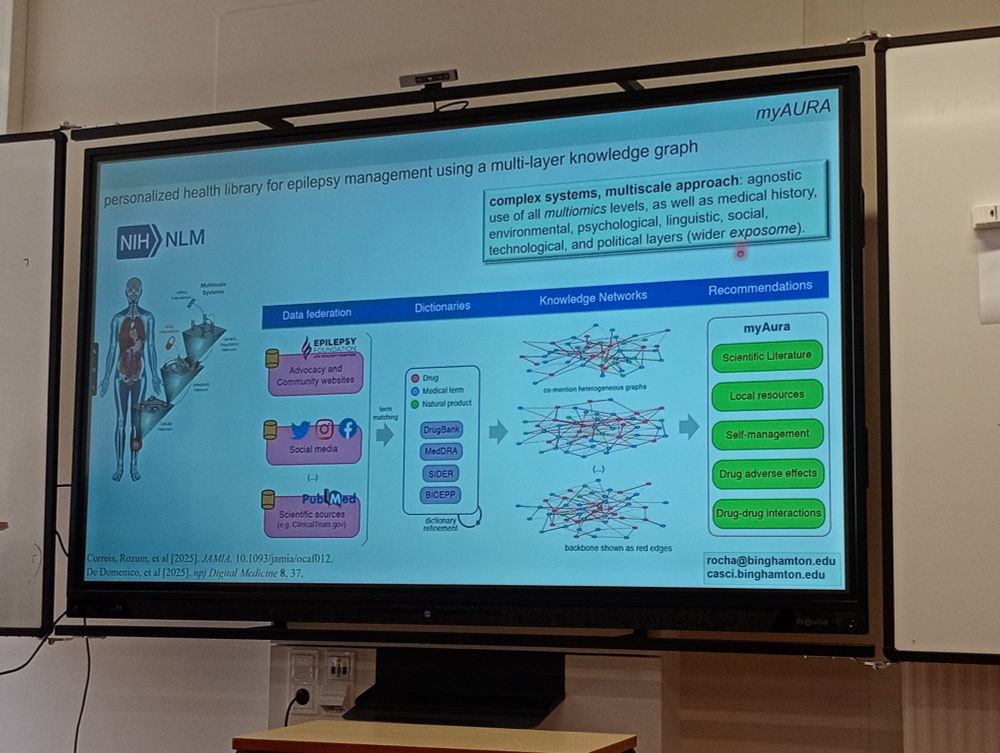
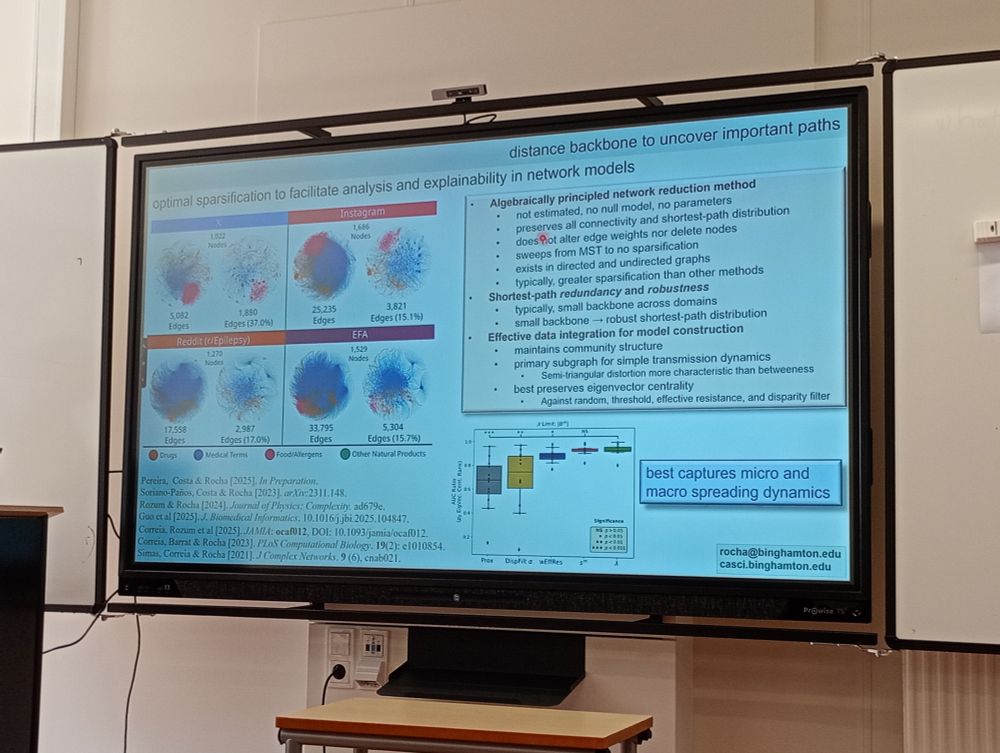
#NetBioMed2025 #NetSci2025

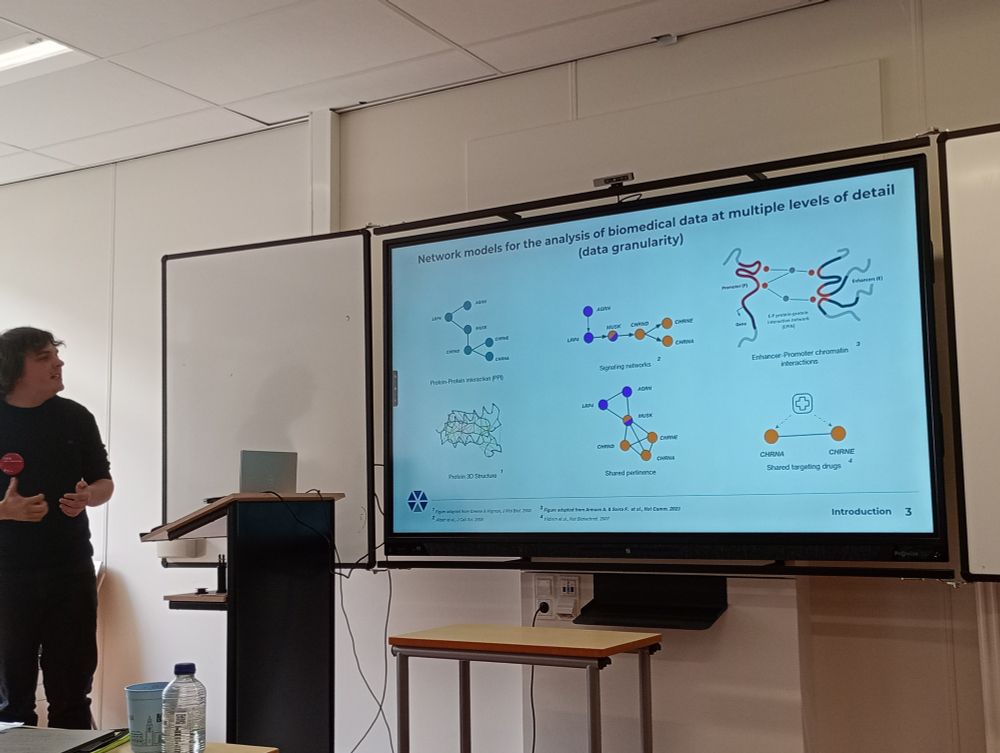

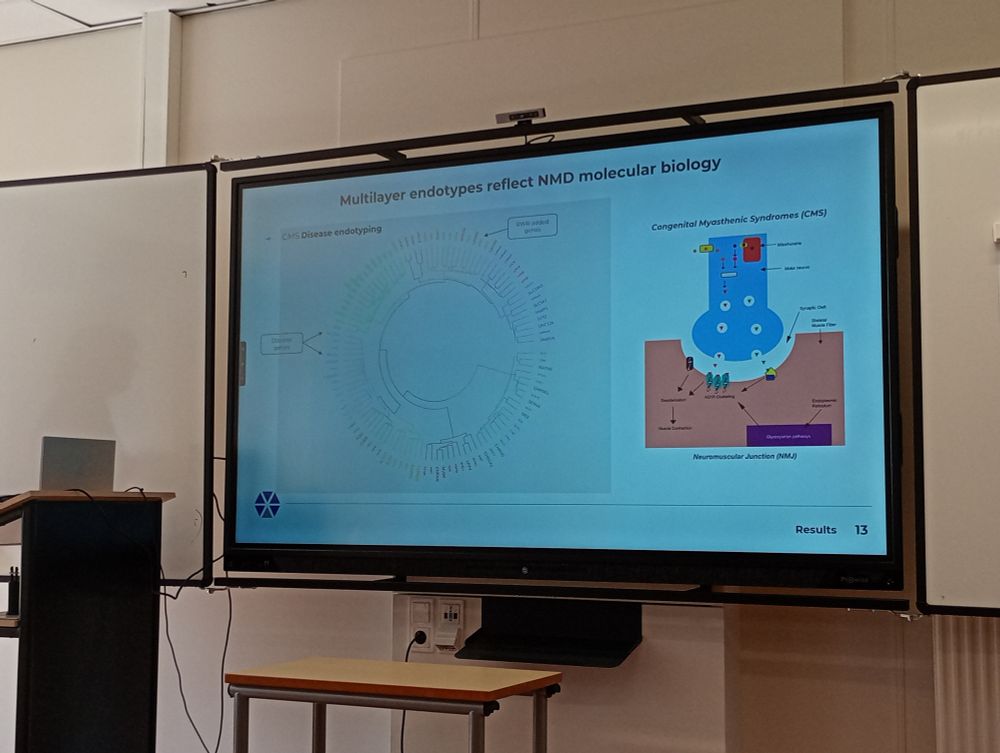
@netsciconf.bsky.social @verapancaldi.bsky.social



@netsciconf.bsky.social @verapancaldi.bsky.social
#NetBioMed2025 #NetSci2025 @verapancaldi.bsky.social

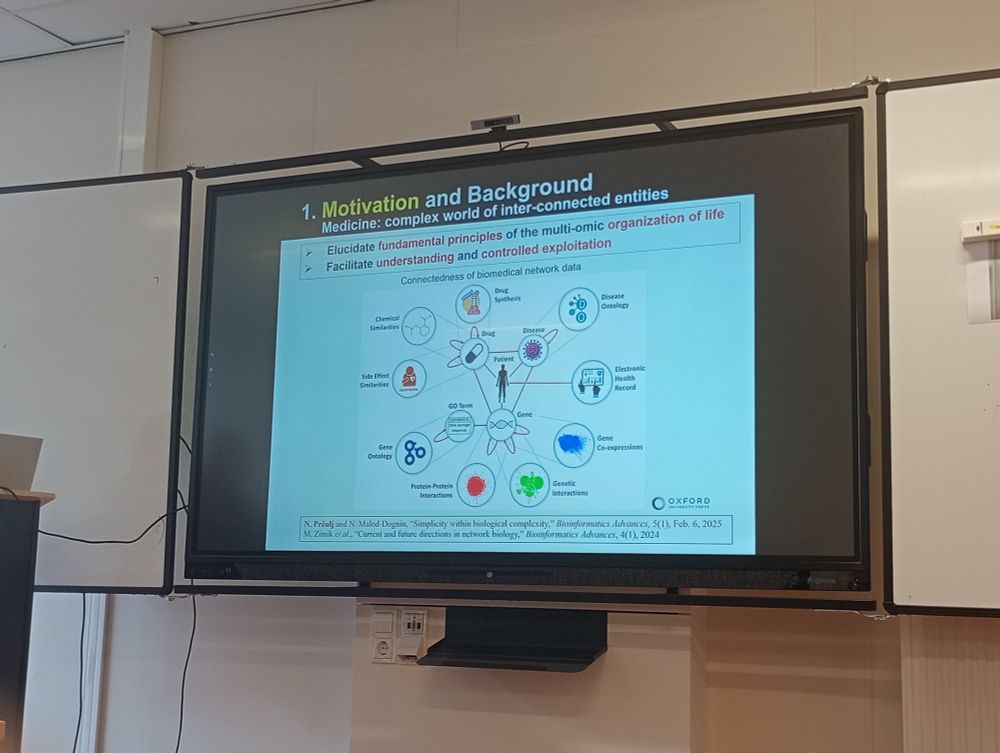

#NetBioMed2025 #NetSci2025 @verapancaldi.bsky.social
To access the work www.nature.com/articles/s41...
Grâce à scPRINT, des chercheurs décryptent les réseaux de gènes qui contrôlent les états cellulaires à partir de millions de données single-cell.
buff.ly/XMcDog0

To access the work www.nature.com/articles/s41...


