clareaulab.com
@fredsteammskcc.bsky.social and
@mskcancercenter.bsky.social! Shout out to the lab for supporting us!

@fredsteammskcc.bsky.social and
@mskcancercenter.bsky.social! Shout out to the lab for supporting us!





My first ultra in ~9 years… Couldn’t have done it without you! Don’s fries hit different after mile 80 lol

My first ultra in ~9 years… Couldn’t have done it without you! Don’s fries hit different after mile 80 lol
Together, we ran 303.3 miles in under 75 hours, burning 50,000+ calories along the way!

Together, we ran 303.3 miles in under 75 hours, burning 50,000+ calories along the way!
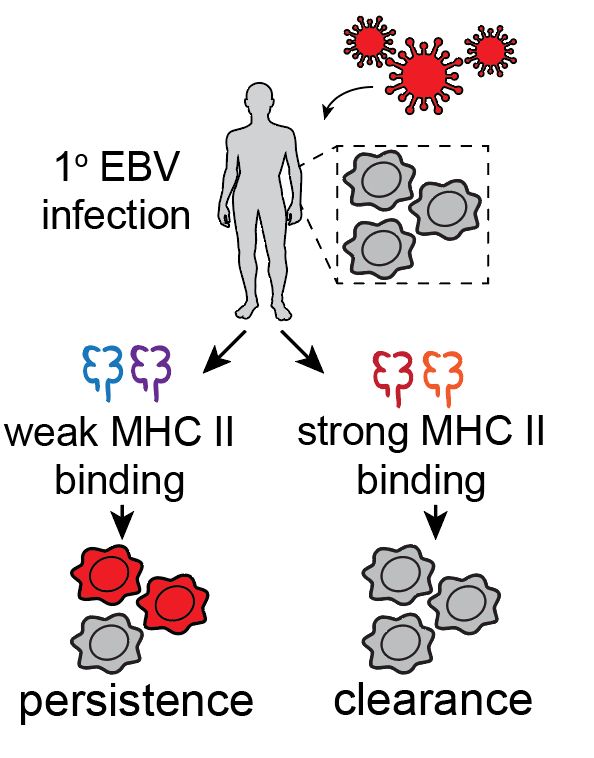
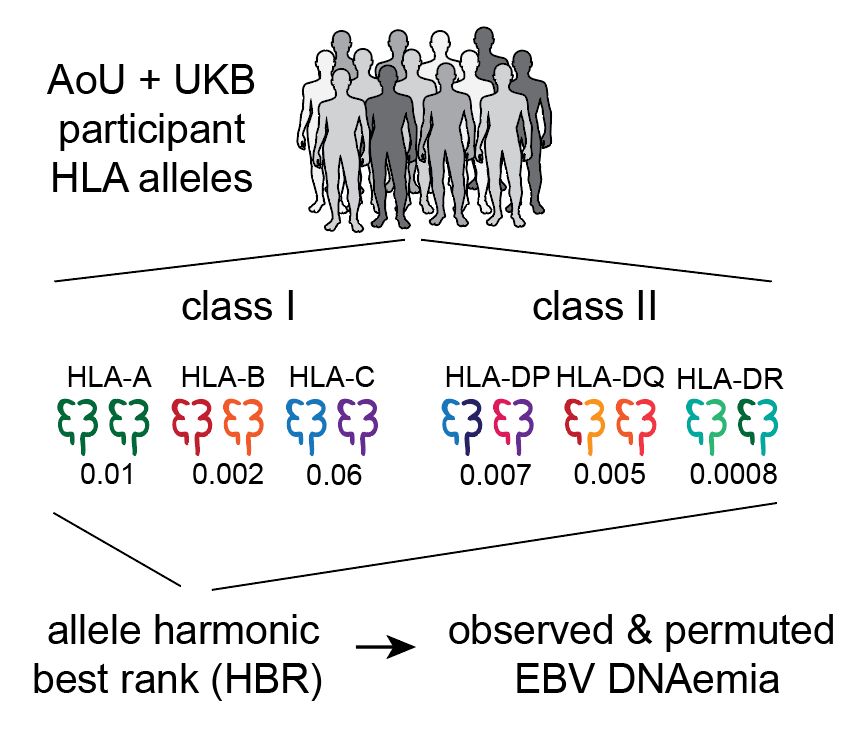
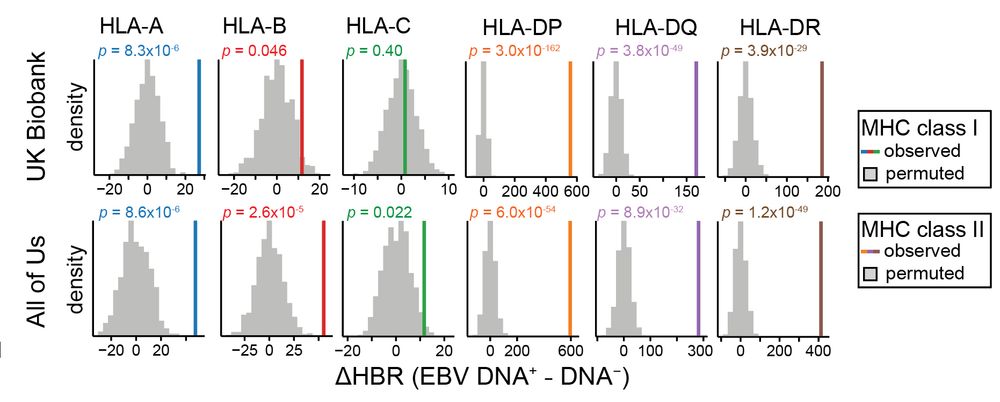

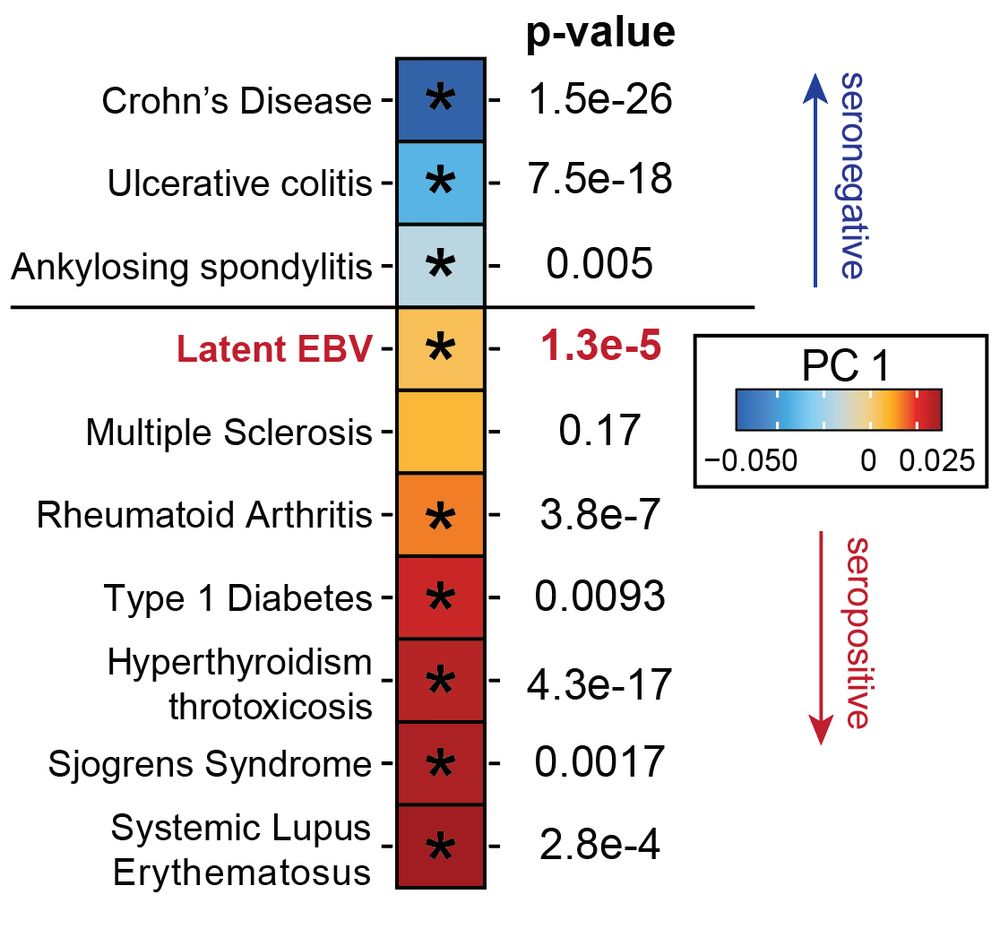
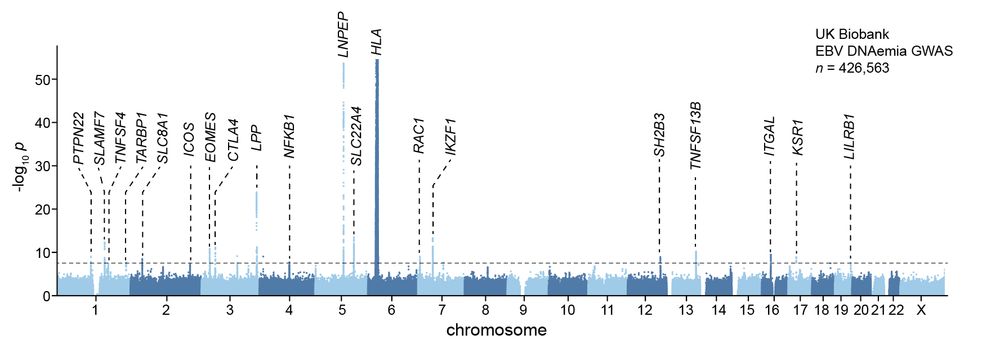
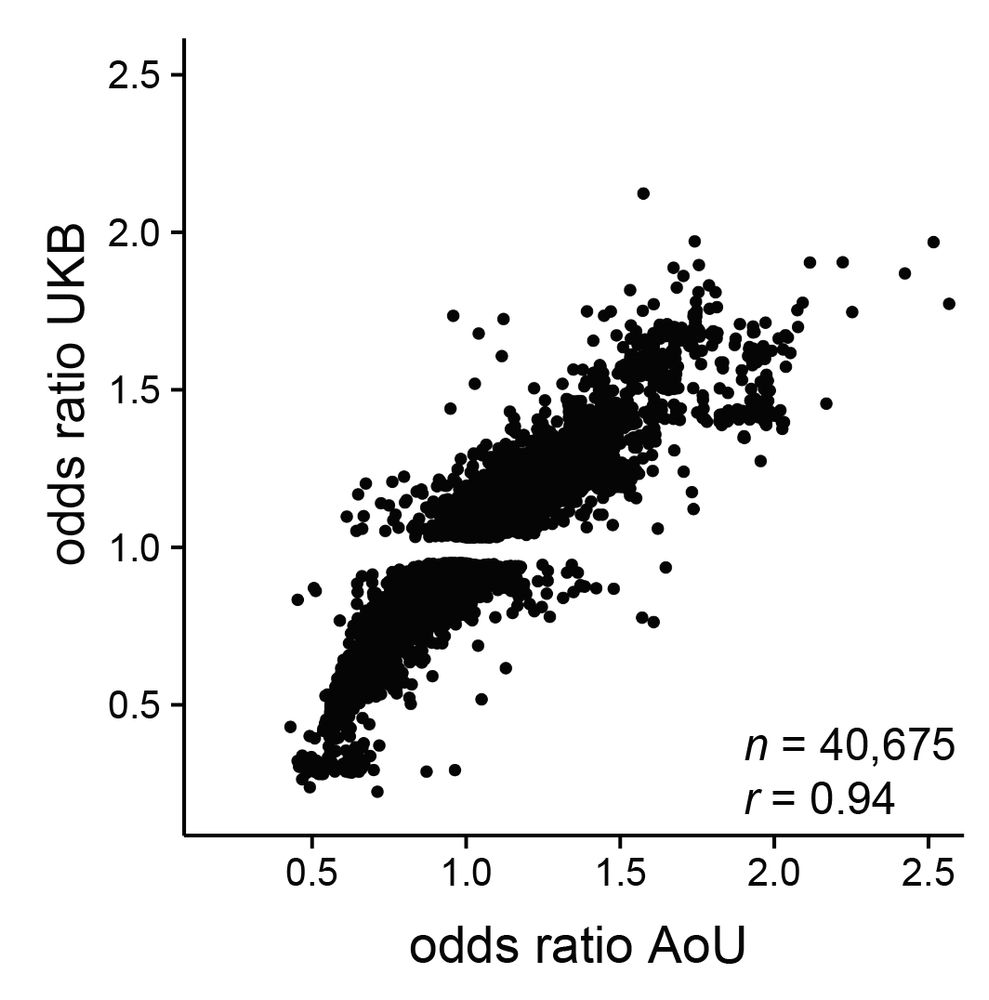
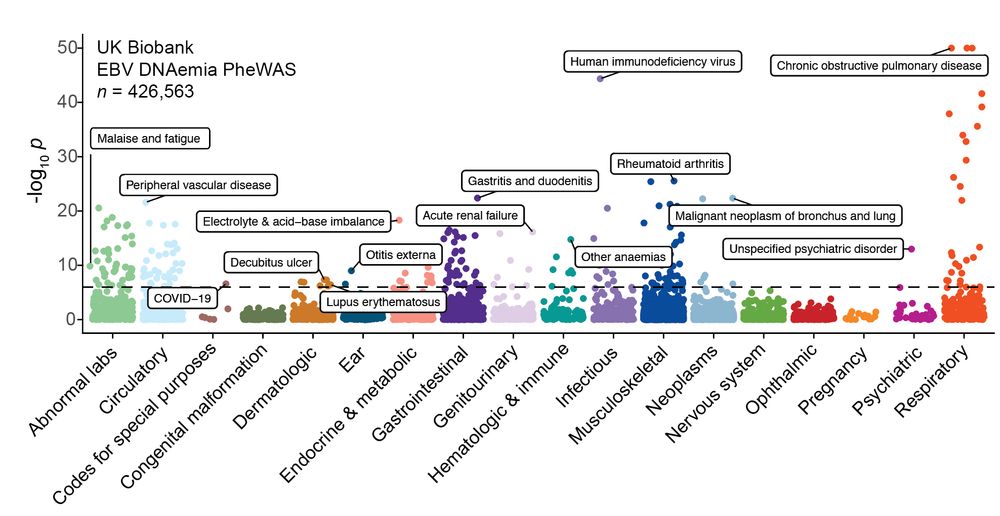
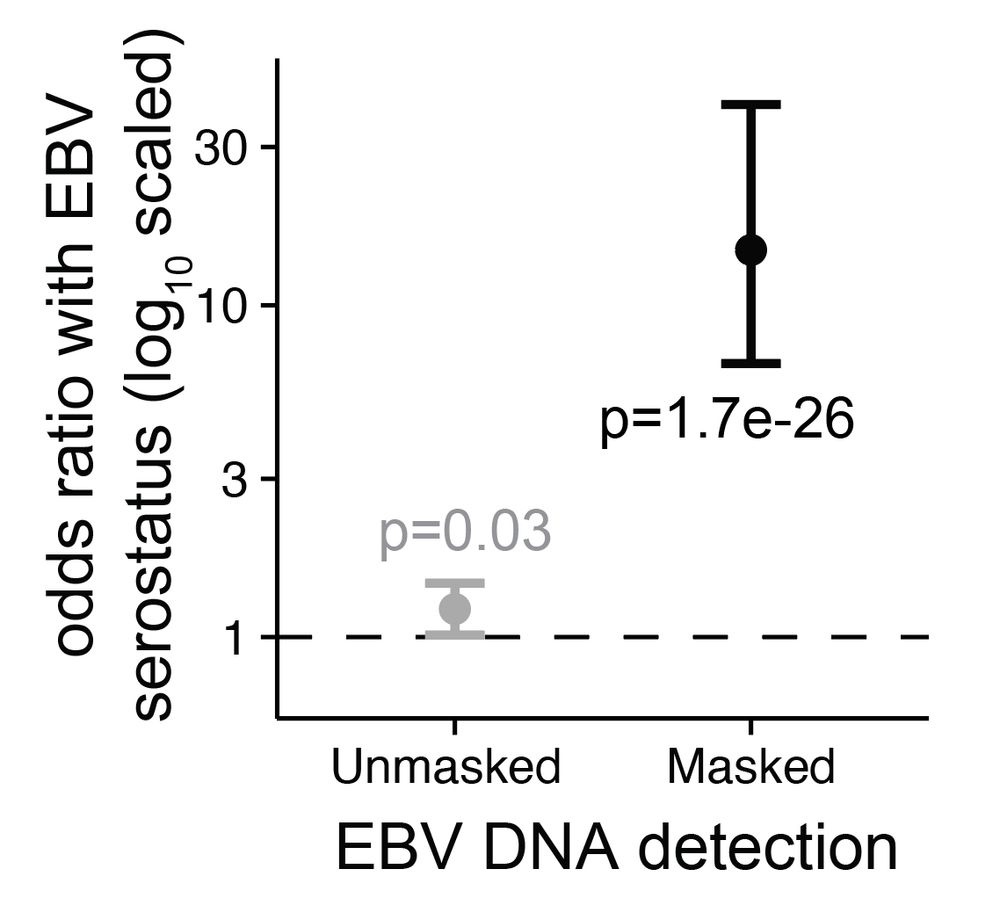
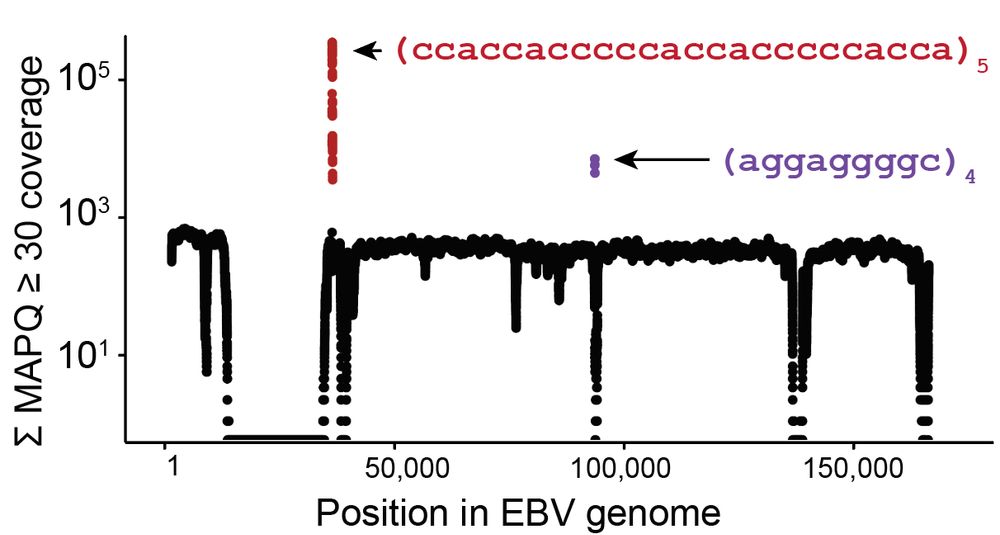
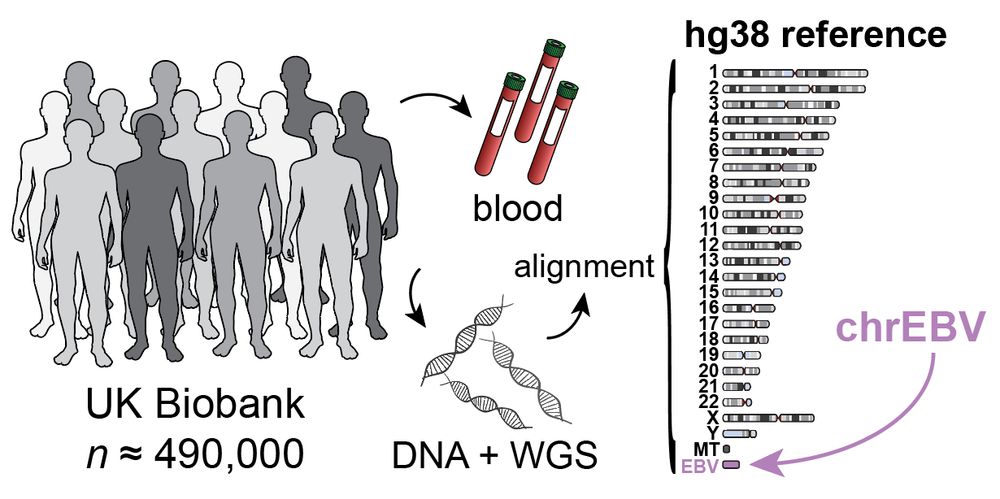
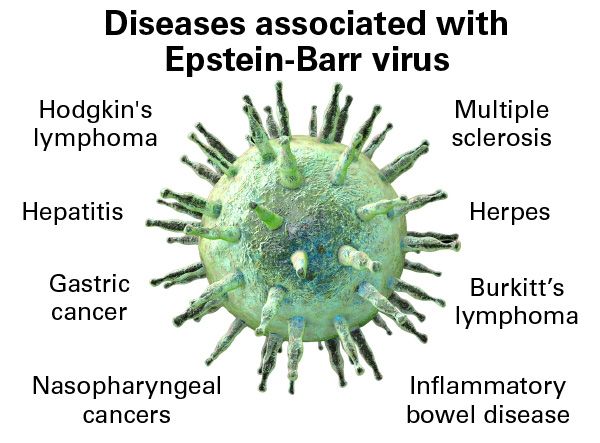
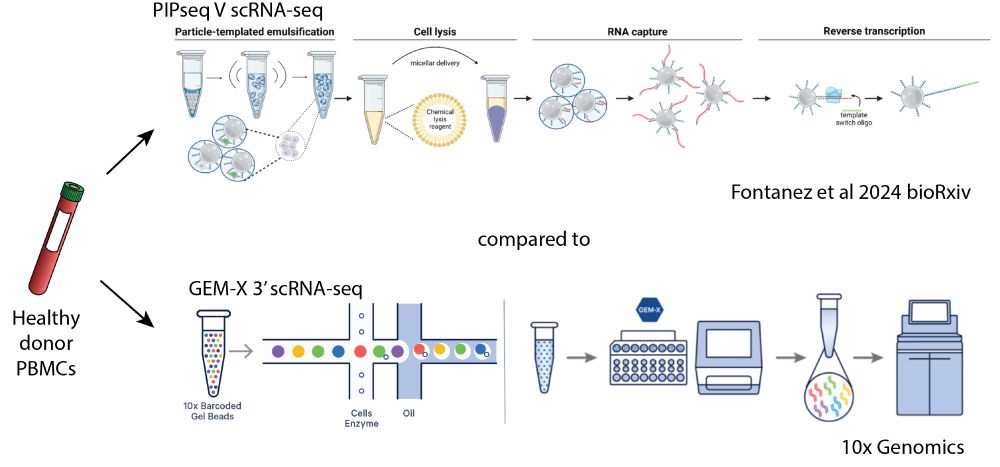
Led by @sherrynyeo.bsky.social, @erinmayc.bsky.social, and friends, we continue our journey to find viral DNA in our favorite place-- the overlooked and discarded reads in existing data! 1/

Led by @sherrynyeo.bsky.social, @erinmayc.bsky.social, and friends, we continue our journey to find viral DNA in our favorite place-- the overlooked and discarded reads in existing data! 1/