🧩 Know of an important motif? Add it to the benchmark!
🏗️ Help improve the pipeline & metrics (sequence-based? Side-chain-level?)
Let’s shape the future of motif scaffolding together!
🧩 Know of an important motif? Add it to the benchmark!
🏗️ Help improve the pipeline & metrics (sequence-based? Side-chain-level?)
Let’s shape the future of motif scaffolding together!
This suggests modern deep learning methods aren’t always better than past methods!

This suggests modern deep learning methods aren’t always better than past methods!
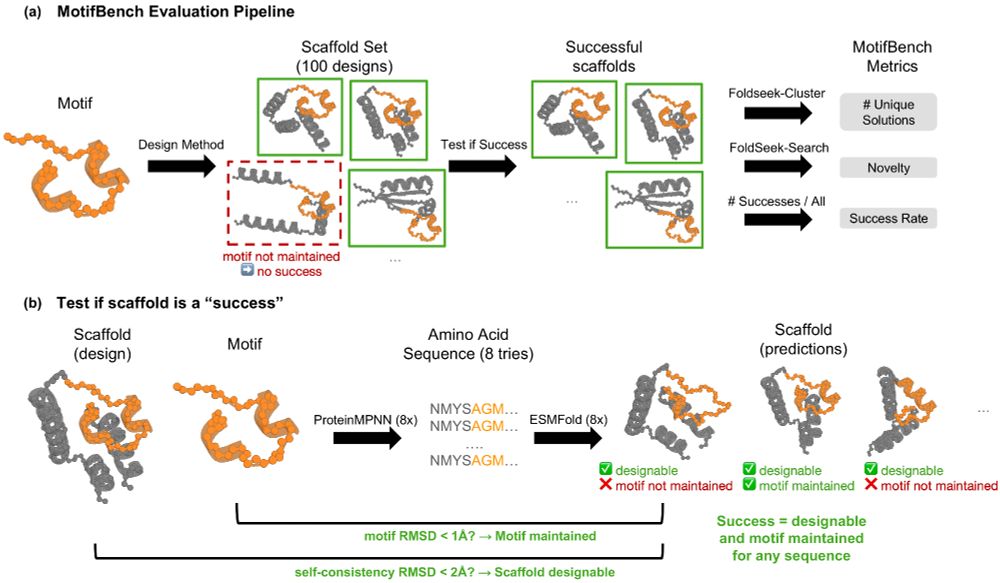
🧪 A standardized evaluation pipeline
🏆 30 challenging motifs as test cases
📊Easy-to-use eval scripts and a leaderboard for method comparison
Now, results can be easily and consistently measured.
🧪 A standardized evaluation pipeline
🏆 30 challenging motifs as test cases
📊Easy-to-use eval scripts and a leaderboard for method comparison
Now, results can be easily and consistently measured.
❌ Current evaluation are inconsistent, and results incomparable
❌ Widely used test cases are too easy
❌ Reproducibility is difficult
That’s where MotifBench comes in.
❌ Current evaluation are inconsistent, and results incomparable
❌ Widely used test cases are too easy
❌ Reproducibility is difficult
That’s where MotifBench comes in.
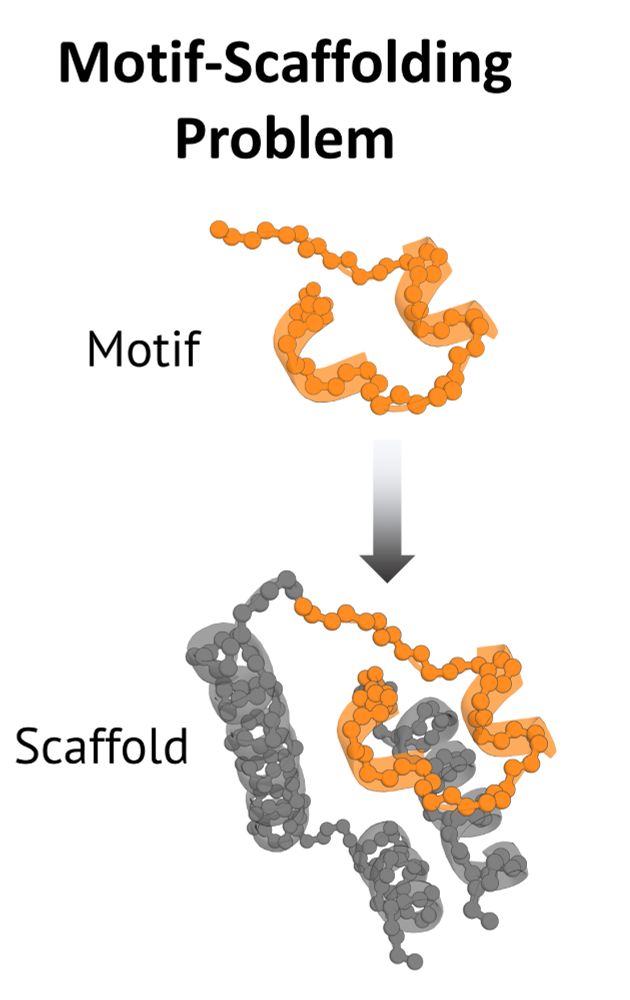
✅ Input: a motif (small functional substructure)
🎯 Goal: find a scaffolds (full proteins) that preserves the motif’s geometry.
But what's the state of methods for this problem?
✅ Input: a motif (small functional substructure)
🎯 Goal: find a scaffolds (full proteins) that preserves the motif’s geometry.
But what's the state of methods for this problem?

