
Site: https://biapyx.github.io/
Doc: https://biapy.readthedocs.io/en/latest/
Code: https://github.com/BiaPyX/BiaPy
Try the tool 👉 imaging.usegalaxy.eu?tool_id=tool...
Contribute your models 🧑🔬 And join the community making bioimage analysis open & reproducible!
#BiaPy #GalaxyProject #BioImageAnalysis #Bioimaging
Try the tool 👉 imaging.usegalaxy.eu?tool_id=tool...
Contribute your models 🧑🔬 And join the community making bioimage analysis open & reproducible!
#BiaPy #GalaxyProject #BioImageAnalysis #Bioimaging
Pretrained models can now be run straight through Galaxy with BiaPy, bringing community AI models to your data in just a few clicks.
#BioImageModelZoo #OpenScience

Pretrained models can now be run straight through Galaxy with BiaPy, bringing community AI models to your data in just a few clicks.
#BioImageModelZoo #OpenScience
That means:
✔️ Step-by-step experiment setup
✔️ Launch training / inference jobs with BiaPy under the hood
✔️ Share & reproduce pipelines the Galaxy way 🌌

That means:
✔️ Step-by-step experiment setup
✔️ Launch training / inference jobs with BiaPy under the hood
✔️ Share & reproduce pipelines the Galaxy way 🌌
Huge thanks and keep it up! 💪

Huge thanks and keep it up! 💪
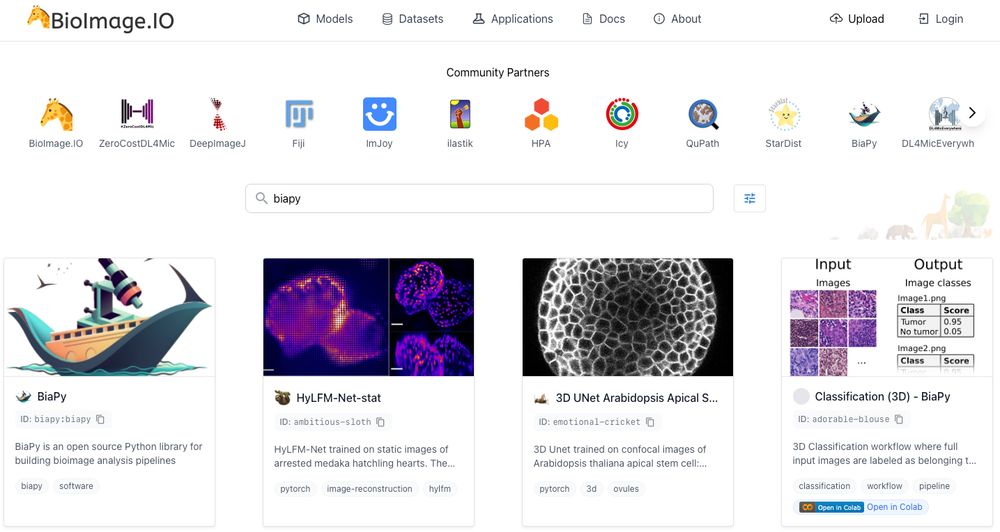
🔗 Have a look at: bioimage.io#/models
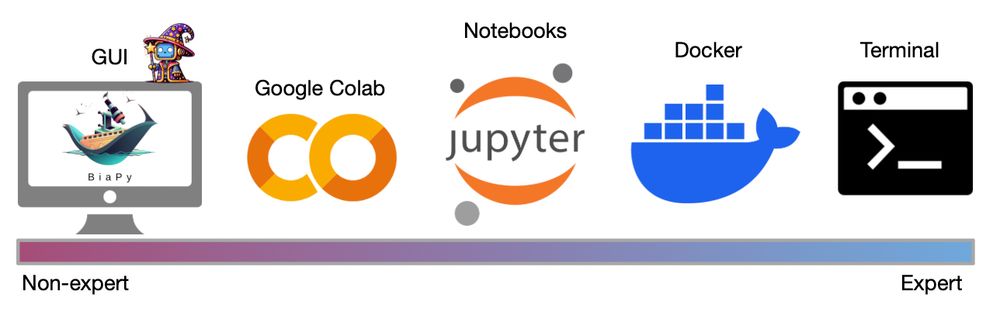
As usual, they are accesible from BiaPy's GUI 🖥️, Colab notebooks 📔 and our API 🤓
🔗 Have a look at: bioimage.io#/models
As usual, they are accesible from BiaPy's GUI 🖥️, Colab notebooks 📔 and our API 🤓
As an example, we can now reuse the predictions of a workflow and execute only the post-processing step with different parameters, which is much faster ⏩
biapy.readthedocs.io/en/latest/ge...
As an example, we can now reuse the predictions of a workflow and execute only the post-processing step with different parameters, which is much faster ⏩
biapy.readthedocs.io/en/latest/ge...
dipc.ehu.eus/en/career/in...

dipc.ehu.eus/en/career/in...
bsky.app/profile/biap...
So no more this 👇
bsky.app/profile/biap...
So no more this 👇


@naturemethods.bsky.social, @ritastrack.bsky.social, the reviewers, and the awesome bioimage analysis community.
We can't wait to see what you build with BiaPy! 🚀🧠
🔗 www.nature.com/articles/s41...
#BiaPy #BioimageAnalysis #DeepLearning
@naturemethods.bsky.social, @ritastrack.bsky.social, the reviewers, and the awesome bioimage analysis community.
We can't wait to see what you build with BiaPy! 🚀🧠
🔗 www.nature.com/articles/s41...
#BiaPy #BioimageAnalysis #DeepLearning
Check out the full documentation and tutorials here:
👉 biapy.readthedocs.io
👉 www.youtube.com/@BiaPyX
Contribute, test, and join the community — BiaPy is open source! (6/7)
#OpenScience #Microscopy #AI
Check out the full documentation and tutorials here:
👉 biapy.readthedocs.io
👉 www.youtube.com/@BiaPyX
Contribute, test, and join the community — BiaPy is open source! (6/7)
#OpenScience #Microscopy #AI
@iarganda.bsky.social and @arratemunoz.bsky.social.
A huge thanks to everyone who contributed! 🌟 (5/7)

@iarganda.bsky.social and @arratemunoz.bsky.social.
A huge thanks to everyone who contributed! 🌟 (5/7)
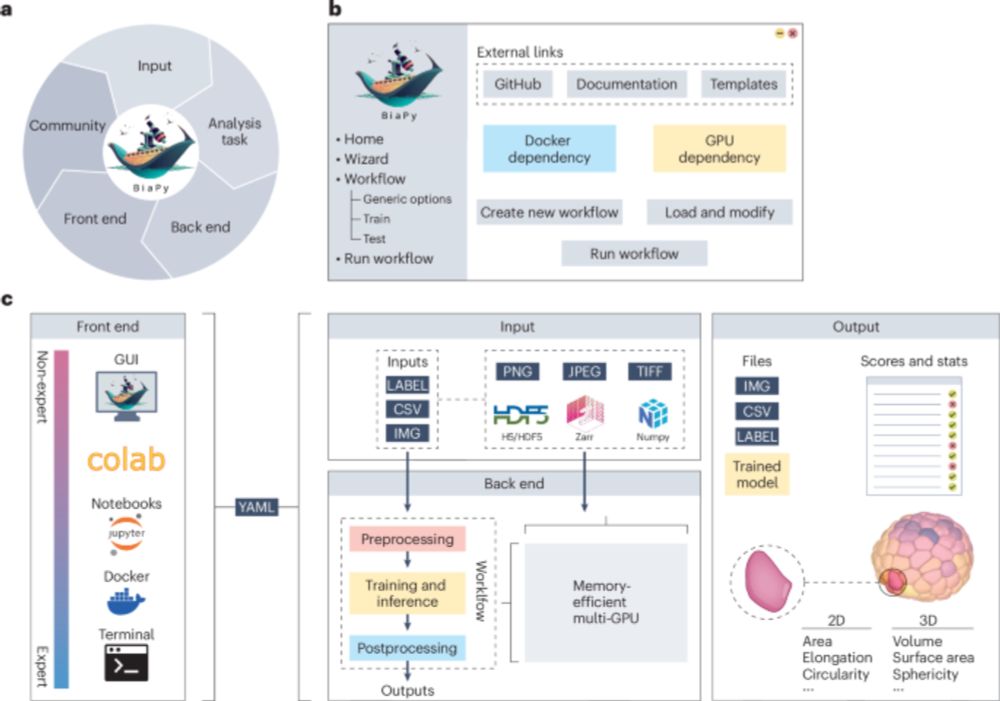
• Full multi-GPU support for training and inference
• Efficient memory management for very large datasets (MBs → TBs)
• Single-container installation via Docker
• Integration with @BioImageModelZoo for easy model sharing and reuse!
#DeepLearning (4/7)
• Full multi-GPU support for training and inference
• Efficient memory management for very large datasets (MBs → TBs)
• Single-container installation via Docker
• Integration with @BioImageModelZoo for easy model sharing and reuse!
#DeepLearning (4/7)
✅ Instance and semantic segmentation
✅ Object detection
✅ Image classification
✅ Image denoising & super-resolution
✅ Self-supervised learning
✅ Image-to-image translation
From 2D images to full 3D brain scans! 🧠 (3/7)
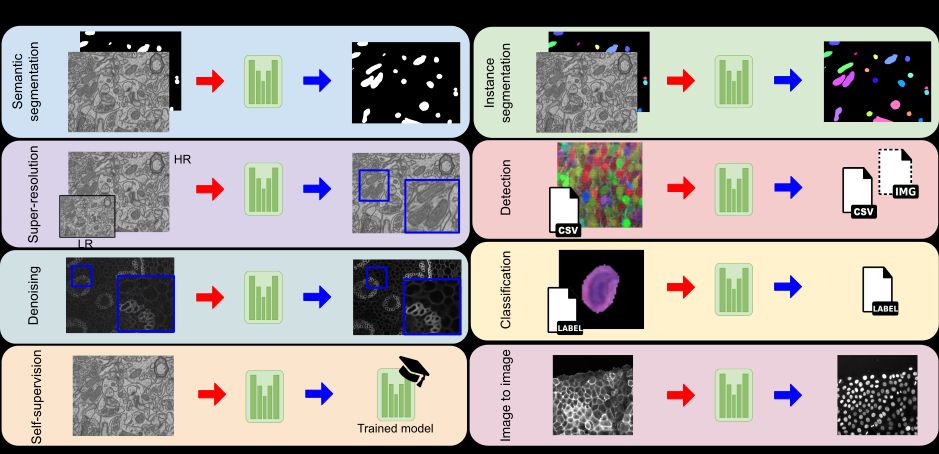
✅ Instance and semantic segmentation
✅ Object detection
✅ Image classification
✅ Image denoising & super-resolution
✅ Self-supervised learning
✅ Image-to-image translation
From 2D images to full 3D brain scans! 🧠 (3/7)


