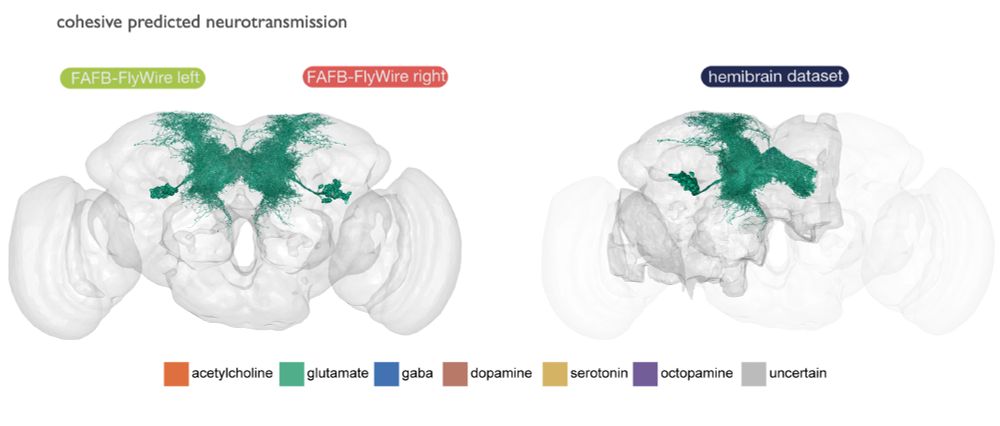
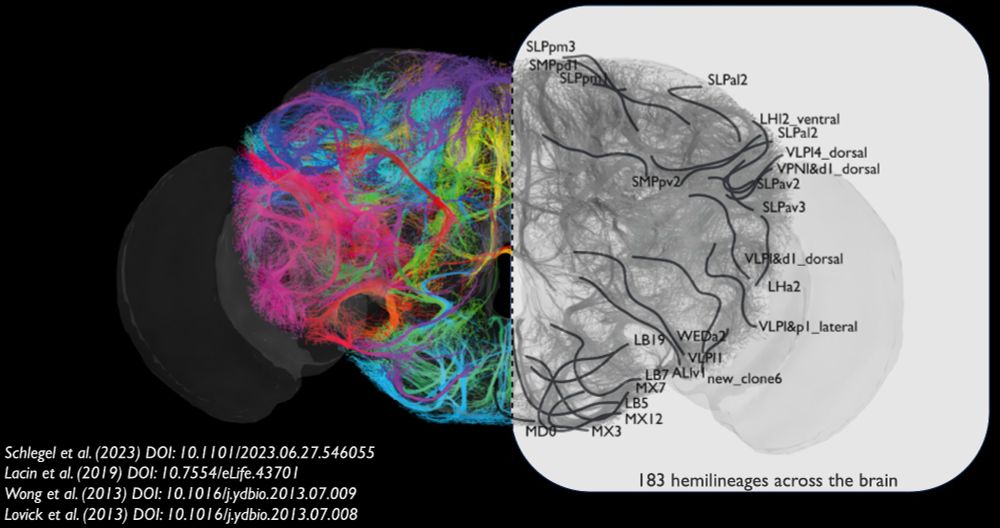
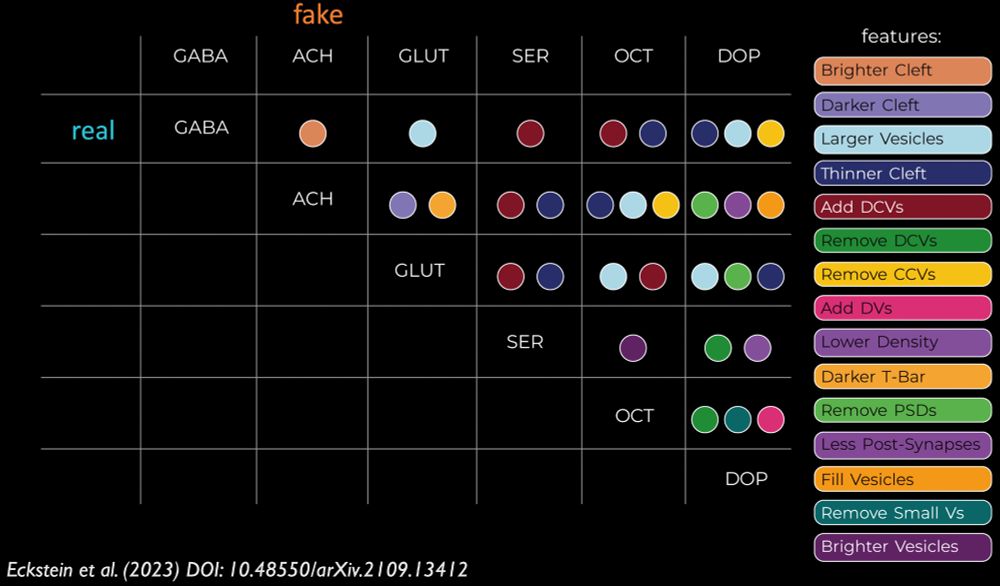
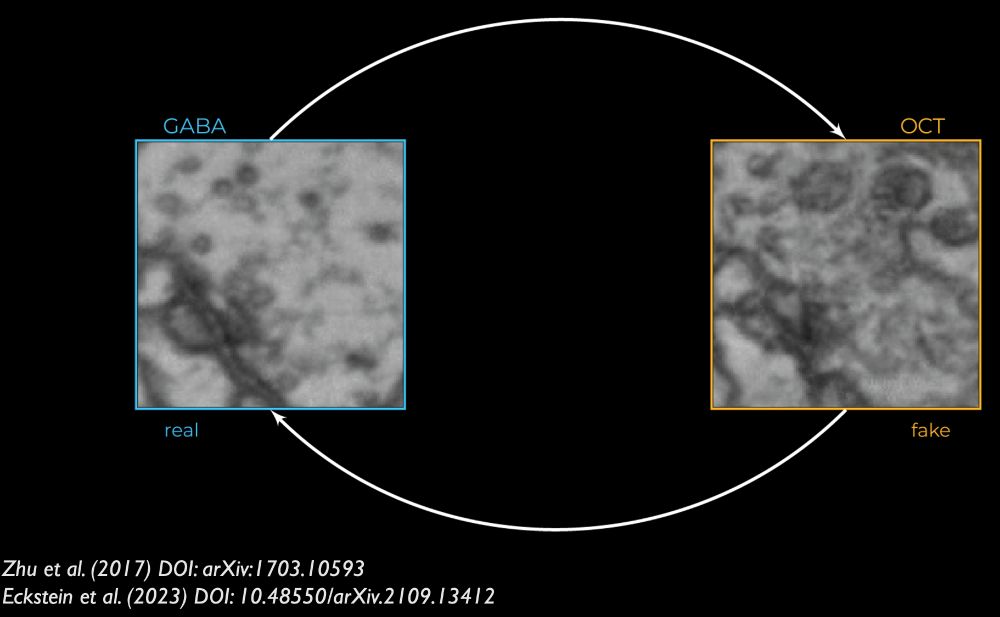
Details: github.com/jasper-tms/t...

Details: github.com/jasper-tms/t...
(graphics: @mottcallie.bsky.social, tool: Arie Matisliah)
Vector graphics! github.com/wilson-lab/s...
Effector cells in 3D! ng.banc.community/2025a/effere...

(graphics: @mottcallie.bsky.social, tool: Arie Matisliah)
Vector graphics! github.com/wilson-lab/s...
Effector cells in 3D! ng.banc.community/2025a/effere...

Different from prior connectomes - it is brain + cord (think spinal cord)
We use it to ‘embody’ the system and find it resembles ‘subsumption architecture’ doi.org/10.1101/2025...
Different from prior connectomes - it is brain + cord (think spinal cord)
We use it to ‘embody’ the system and find it resembles ‘subsumption architecture’ doi.org/10.1101/2025...