PhD from Schatz Lab @ JHU. Previously: CS @ Brown. He/His/Him. #YNWA 🍉
Thank you to @mikeschatz.bsky.social, @rajivmccoy.bsky.social and @aabiddanda.bsky.social for all their work on this.
🧵 A thread on the key results and takeaways from our work:


Papers biorxiv.org/content/10.1101/2025.05.20.654611 & doi.org/10.1186/s13059-025-03644-0
Code: github.com/vikshiv/mume...
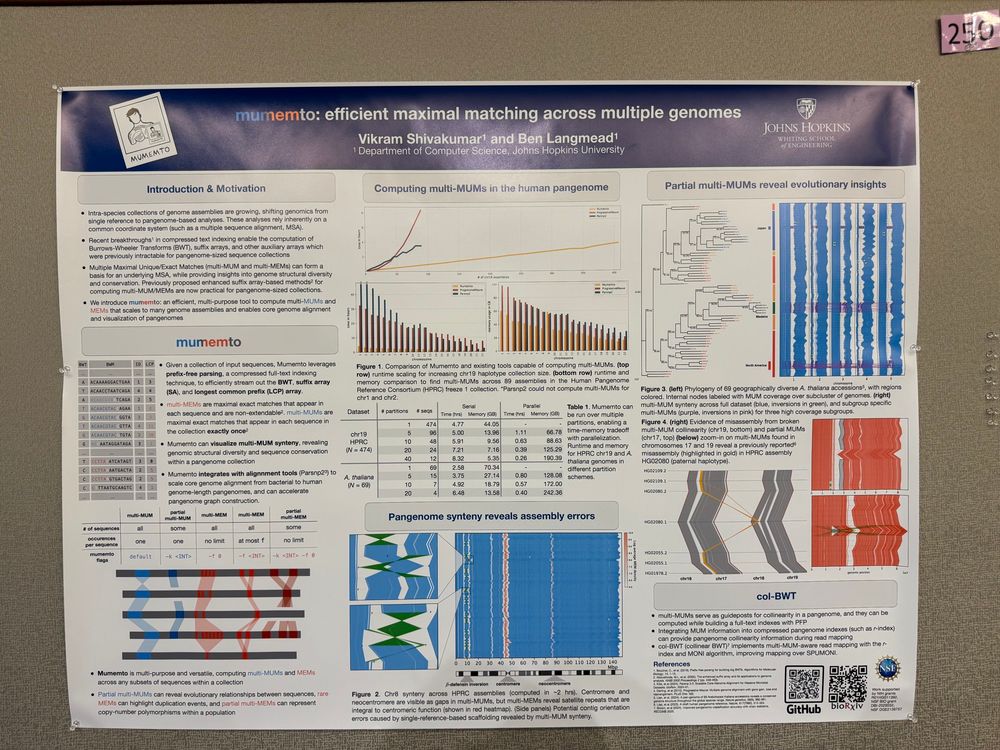
Very efficient pangenome visualization tool, revealing synteny and variations!



Papers biorxiv.org/content/10.1101/2025.05.20.654611 & doi.org/10.1186/s13059-025-03644-0
Code: github.com/vikshiv/mume...
Very efficient pangenome visualization tool, revealing synteny and variations!
Taken from this resource from my alma mater: costsofwar.watson.brown.edu
(Specific page is: costsofwar.watson.brown.edu/costs/human/...)

Taken from this resource from my alma mater: costsofwar.watson.brown.edu
(Specific page is: costsofwar.watson.brown.edu/costs/human/...)

Wish it was made clear in the initial “proclamation”, before we spent the entire day panicking while trying to figure out a way to get a friend back to the US before midnight.

Wish it was made clear in the initial “proclamation”, before we spent the entire day panicking while trying to figure out a way to get a friend back to the US before midnight.
So, so many people I know and love are going to find it impossible to stay and work in the US, and it makes it almost impossible for people like me to stay and work here in the longer term, no matter how qualified we are.
So, so many people I know and love are going to find it impossible to stay and work in the US, and it makes it almost impossible for people like me to stay and work here in the longer term, no matter how qualified we are.
lh3.github.io/2025/09/11/a...

lh3.github.io/2025/09/11/a...
Spineless and shameful.

Spineless and shameful.
Mumemto scales to the new HPRC v2 release and beyond, and can merge in future assemblies without any recomputation! 1/n

Mumemto scales to the new HPRC v2 release and beyond, and can merge in future assemblies without any recomputation! 1/n
Come and watch my friend Sara defend her PhD!

Come and watch my friend Sara defend her PhD!
Thank you to @mikeschatz.bsky.social, @rajivmccoy.bsky.social and @aabiddanda.bsky.social for all their work on this.
🧵 A thread on the key results and takeaways from our work:
Thank you to @mikeschatz.bsky.social, @rajivmccoy.bsky.social and @aabiddanda.bsky.social for all their work on this.
🧵 A thread on the key results and takeaways from our work:
Thank you to @mikeschatz.bsky.social, @rajivmccoy.bsky.social and @aabiddanda.bsky.social for all their work on this.
🧵 A thread on the key results and takeaways from our work:
Thank you to @mikeschatz.bsky.social, @rajivmccoy.bsky.social and @aabiddanda.bsky.social for all their work on this.
🧵 A thread on the key results and takeaways from our work:
I'm on the job market this summer, so please send any interesting opportunities my way 😃
I'm on the job market this summer, so please send any interesting opportunities my way 😃
I’m going to be defending my thesis on Wednesday, so I thought this was as good a time as any to introduce myself and my work.
I’m Arun Das, I’m a PhD student in Schatz Lab @ JHU, and my work broadly focuses on algorithms to improve accessibility and representation in genomics.
I’m going to be defending my thesis on Wednesday, so I thought this was as good a time as any to introduce myself and my work.
I’m Arun Das, I’m a PhD student in Schatz Lab @ JHU, and my work broadly focuses on algorithms to improve accessibility and representation in genomics.


