

---
#proteomics #prot-paper

---
#proteomics #prot-paper
---
#proteomics #prot-paper
---
#proteomics #prot-paper
www.biorxiv.org/content/10.1...
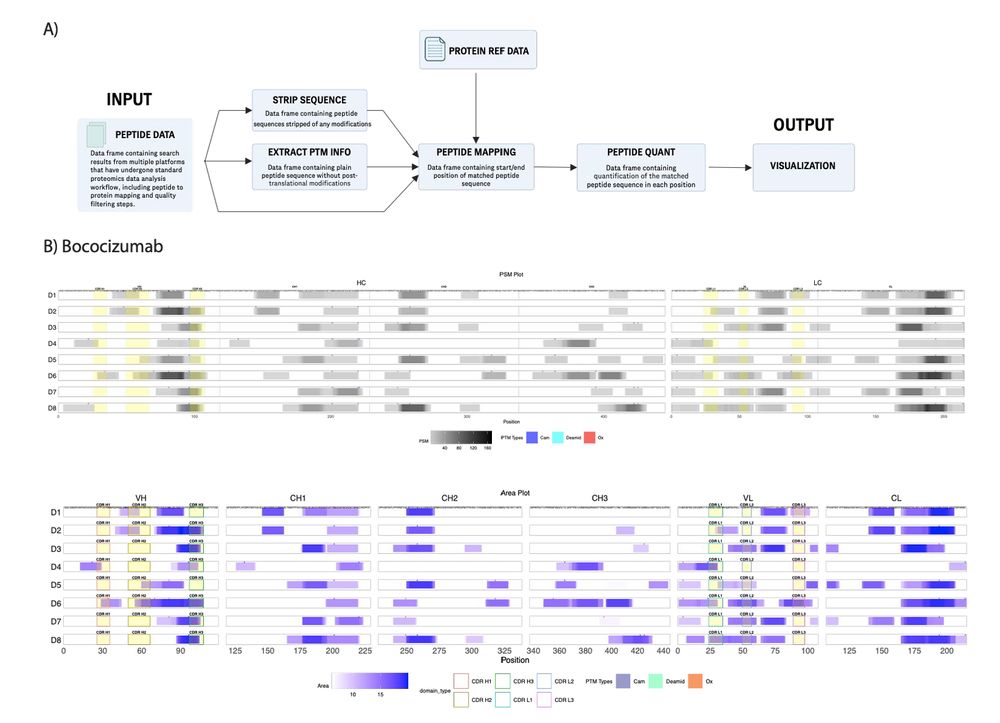
This preprint describes our scalable, mass spec-based method for measuring which (and how many of them) proteins are interacting with DNA.
1/?

www.biorxiv.org/content/10.1...
This preprint describes our scalable, mass spec-based method for measuring which (and how many of them) proteins are interacting with DNA.
1/?
www.verwaltung.uni-halle.de/dezern3/Auss...
#Academicsky
#Chemsky
#teammassspec
www.verwaltung.uni-halle.de/dezern3/Auss...
#Academicsky
#Chemsky
#teammassspec
www.nature.com/articles/s41...
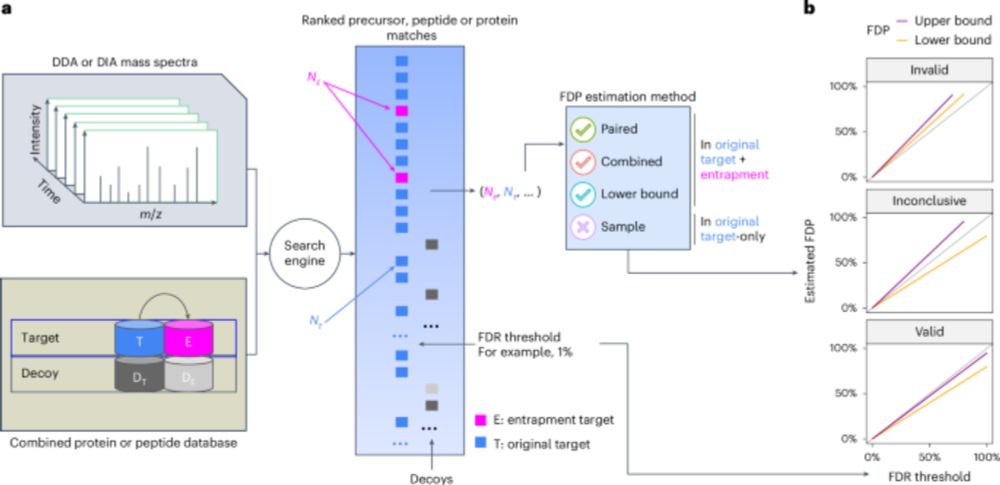
www.nature.com/articles/s41...



The applications for the Empowerment Award for Q3 2025 will open soon 🏆
You can find more information here ➡️ femalesinms.com/empowerment/

The applications for the Empowerment Award for Q3 2025 will open soon 🏆
You can find more information here ➡️ femalesinms.com/empowerment/
Heterobifunctional by warhead, K-K reactive, and partially tris tolerant.
I believe that this could be made into a cleavable version by reducing compound 1b to a primary amine and reacting with DSC to make an NHS-carbamate instead of an NHS-ester. Great work.

Heterobifunctional by warhead, K-K reactive, and partially tris tolerant.
I believe that this could be made into a cleavable version by reducing compound 1b to a primary amine and reacting with DSC to make an NHS-carbamate instead of an NHS-ester. Great work.
📢 Call for Applications 📢
Don’t miss this opportunity!
Applications are open from *May 12th to 23rd* 📧
hispanicsandlatinxinms.org/asms-2025-ea...
#ASMS2025 #MSCommunity

📢 Call for Applications 📢
Don’t miss this opportunity!
Applications are open from *May 12th to 23rd* 📧
hispanicsandlatinxinms.org/asms-2025-ea...
#ASMS2025 #MSCommunity

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
@standupforscience.bsky.social
#supportscience
#saveNIH
@standupforscience.bsky.social
#supportscience
#saveNIH
Thinking of all of the NIH scientists right now.




Thinking of all of the NIH scientists right now.








