Previously PhD @ Sebé-Perdós lab @crg.eu
Interested in regulatory genomics, evolution, machine learning, and especially the combination of all of the above.
https://anamaria.elek.hr/
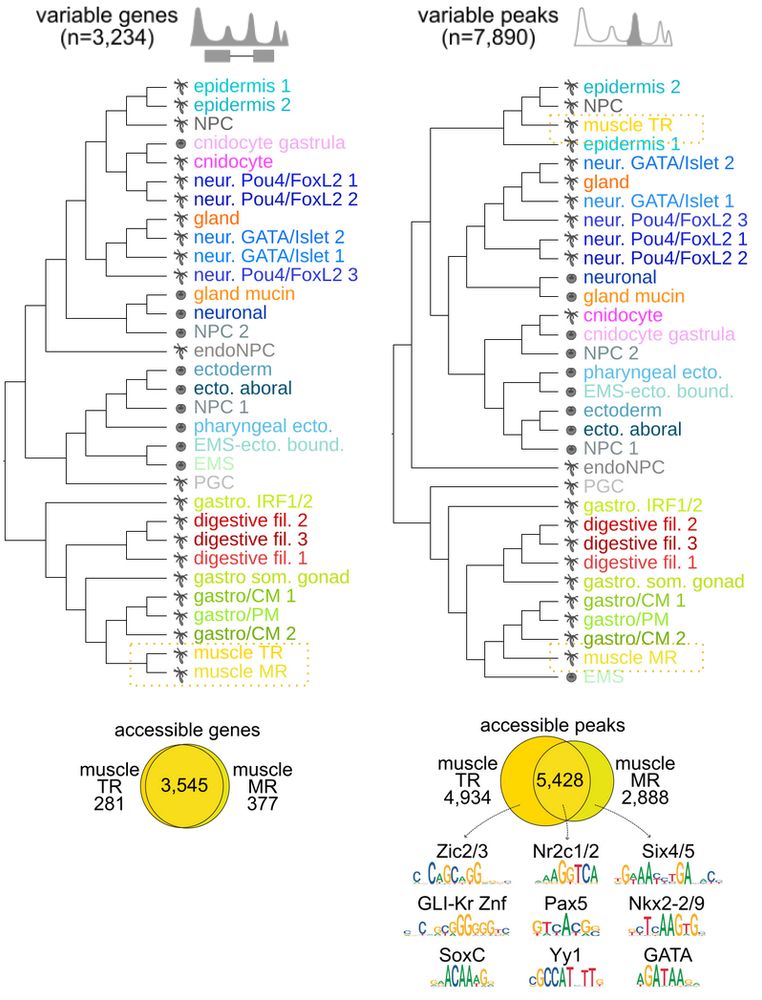
EMS and pharyngeal derivatives, and Pou4/FoxL2 neurons with ectodermal derivatives.
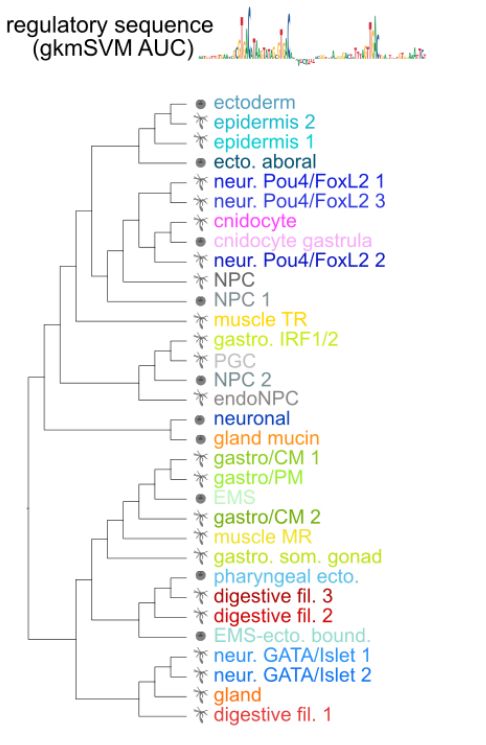
EMS and pharyngeal derivatives, and Pou4/FoxL2 neurons with ectodermal derivatives.