Only real change was N:T271S in one of the two. (A29086T was inherited, but C->G muts like this are very rare.)

Only real change was N:T271S in one of the two. (A29086T was inherited, but C->G muts like this are very rare.)
🎃
💠NB.1.8.1 (Nimbus) 41.9%
💠XFG (Stratus) 23.0%
💠PE.1.4 17.2%
💠MC.10.2.1 9.6%
💠BA.3.2 3.7%
45% of NZ covered by testing:
💠Auckland
💠Rotorua
💠Wellington
💠Christchurch
💠Dunedin
💠Queenstown
poops.nz
2/
🎃
This time from Bristol County, RI on Nov 11. It was about a third of the seqs.
I wonder if it was someone who flew back from Australia through SF?
@ryanhisner.bsky.social @snpoehlm.bsky.social @rajlabn.bsky.social
This time from Bristol County, RI on Nov 11. It was about a third of the seqs.
I wonder if it was someone who flew back from Australia through SF?
@ryanhisner.bsky.social @snpoehlm.bsky.social @rajlabn.bsky.social
The authors were kind enough to mention me and the Variant Hunters Ryan Hisner and Federico Gueli.
www.thelancet.com/journals/lan...
🧵

The authors were kind enough to mention me and the Variant Hunters Ryan Hisner and Federico Gueli.
www.thelancet.com/journals/lan...
🧵
It raises a broader question: Can cryptic wastewater-like lineages transmit?
YES
We knew it happened once. Now we know it's happened at least twice. The results in both cases were not pretty. 1/15
Remember the NJ crytic lineage?
I posted 18 months ago that the Spike was too divergent to predict ACE2 binding, and asked if someone else could figure it out.
Some colleagues took me up on it.
Guess what they found?
1/
It raises a broader question: Can cryptic wastewater-like lineages transmit?
YES
We knew it happened once. Now we know it's happened at least twice. The results in both cases were not pretty. 1/15
nextstrain.org/blog/2025-11...

nextstrain.org/blog/2025-11...
Two BA.3.2 sequences (of 67 total) showed up from New South Wales today, as BA.3.2 continues its creeping geographical spread.
Two BA.3.2 sequences (of 67 total) showed up from New South Wales today, as BA.3.2 continues its creeping geographical spread.
I estimate ~300 BA.3.2.* infections in Perth for the latest week, and ~6,000 across the 12 weeks since BA.3.2.* was first detected.
#COVID19 #SARSCoV2 #BA_3_2 #Australia #WA #Perth
🧵

In this latest batch, there's another furin-cleavage site (FCS)-adjacent ∆QT sequence. It's very closely related to the first one, so there's now no doubt that the deletion is real. 1/3

In this latest batch, there's another furin-cleavage site (FCS)-adjacent ∆QT sequence. It's very closely related to the first one, so there's now no doubt that the deletion is real. 1/3


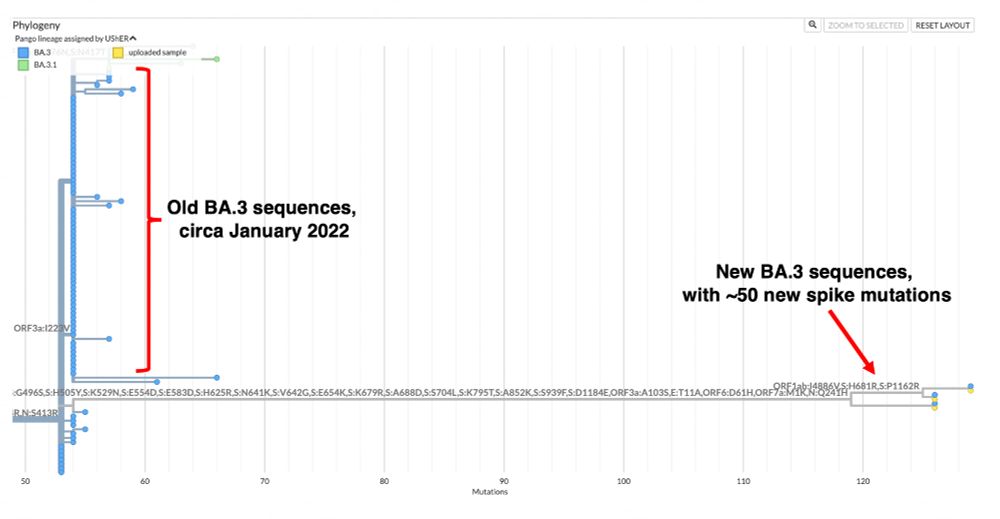
After three years, BA.3 is back.
And it is transmitting.
Who saw this coming?
1/13
He has that power because he conned rich & powerful people into giving it to him. Enough.
This means all public tools/webapps of GISAID data (all the ones you've been used to seeing thru the pandemic, as far as we can tell) are prohibited.
The file allowed this. Cut that - cut off all tools the public & others were using.
He has that power because he conned rich & powerful people into giving it to him. Enough.
🙃

Updating a file and giving access to Nextstrain & Cov-Spectrum does not require extensive resources, so the official justification is a lie. There has to be an ulterior motive here.
Updating a file and giving access to Nextstrain & Cov-Spectrum does not require extensive resources, so the official justification is a lie. There has to be an ulterior motive here.
Same branch as recent BA.3.2.2 from Germany & Slovenia.
It has a few errors (S:ins214:ASDT is misread & ORF1a:E4388K is an artifact). Ignoring those, the one notable new mutation is N:N126K.

Same branch as recent BA.3.2.2 from Germany & Slovenia.
It has a few errors (S:ins214:ASDT is misread & ORF1a:E4388K is an artifact). Ignoring those, the one notable new mutation is N:N126K.
www.youtube.com/watch?v=FuzT...

www.youtube.com/watch?v=FuzT...
BA.3.2 detected in Aotearoa New Zealand wastewater
3.2% for the week to 5 October
BA.3.2 is not shown in the fortnight to 19 Oct. However, variants with a national percentage of < 1% are not included so BA.3.2 is likely to still be circulating

This step is essential for efficient infection of airway epithelial cells.
This step is essential for efficient infection of airway epithelial cells.

The most recent molnupiravir sequences have indeed been from Australia, with the MOV stats for the most recent being pictured below.
Once again, we see strong evidence that MOV-induced mutations are being positively selected. 1/4

The most recent molnupiravir sequences have indeed been from Australia, with the MOV stats for the most recent being pictured below.
Once again, we see strong evidence that MOV-induced mutations are being positively selected. 1/4
cc @snpoehlm.bsky.social @ryanhisner.bsky.social @josetteschoenma.bsky.social ( i m apparently blocked by X, likely it doesn't like intl law)
One of the two QT repeats appears to have been deleted. I've never seen anything like this before. BA.3.2 is a different beast.

One of the two QT repeats appears to have been deleted. I've never seen anything like this before. BA.3.2 is a different beast.


