Richard Michael
@rdmichael.bsky.social
160 followers
340 following
5 posts
PhD student at University of Copenhagen, @DIKU, @KU_BioML group, DDSA fellow.
Posts
Media
Videos
Starter Packs
Reposted by Richard Michael
Reposted by Richard Michael
Reposted by Richard Michael
Reposted by Richard Michael
Reposted by Richard Michael
Frank Noe
@franknoe.bsky.social
· Feb 19
Reposted by Richard Michael
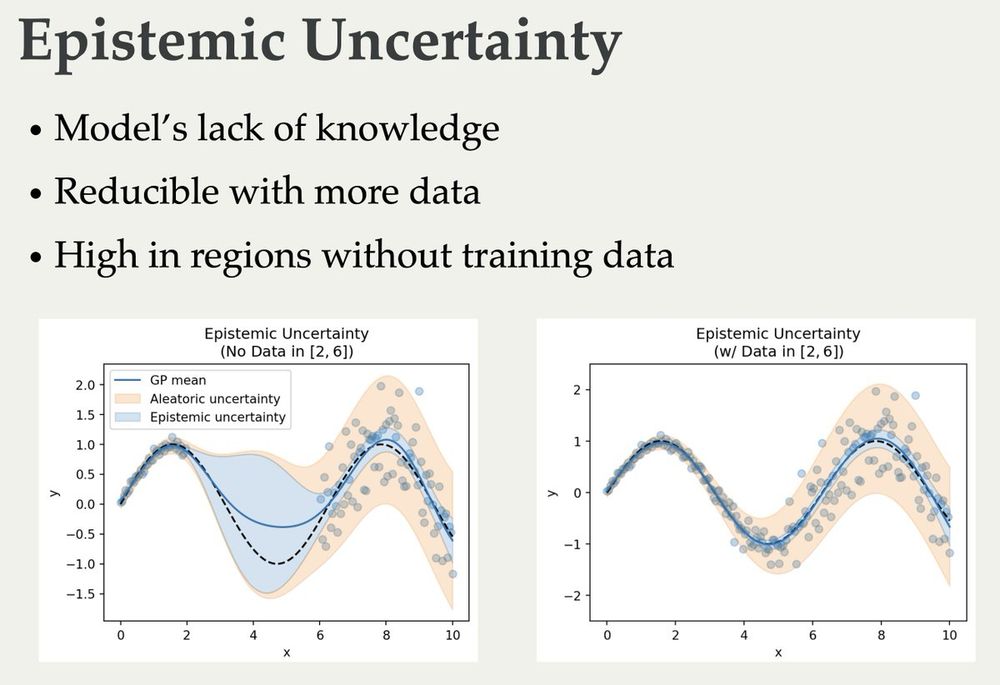
Richard Michael
@rdmichael.bsky.social
· Dec 12
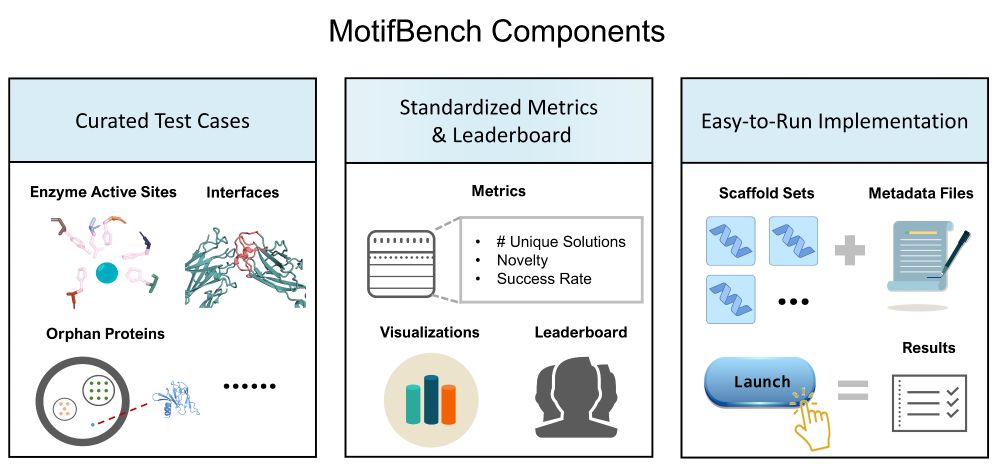
Richard Michael
@rdmichael.bsky.social
· Dec 12
GitHub - MachineLearningLifeScience/poli-baselines: A collection of objective functions and black box optimization algorithms related to proteins and small molecules
A collection of objective functions and black box optimization algorithms related to proteins and small molecules - MachineLearningLifeScience/poli-baselines
github.com
Reposted by Richard Michael