
The peptides suggested another 35 potential new coding genes, but the devil is in the detail. Thread shortly:
bmcgenomics.biomedcentral.com/articles/10....

The peptides suggested another 35 potential new coding genes, but the devil is in the detail. Thread shortly:
bmcgenomics.biomedcentral.com/articles/10....

The peptides suggested another 35 potential new coding genes, but the devil is in the detail. Thread shortly:
bmcgenomics.biomedcentral.com/articles/10....
🌡️ 2024 was the hottest year in recorded history.
Read the latest Climate report from @williamripple.bsky.social and Christopher Wolf's team, published in BioScience.
oxford.ly/47i6K5j

🌡️ 2024 was the hottest year in recorded history.
Read the latest Climate report from @williamripple.bsky.social and Christopher Wolf's team, published in BioScience.
oxford.ly/47i6K5j
GENCODE added loss of function genes to this version and we discovered that this caused the TRIFID scores to change, affecting some of the PRINCIPAL:2 -> PRINCIPAL:5 isoforms.
Our fault, we asked for the change!
GENCODE added loss of function genes to this version and we discovered that this caused the TRIFID scores to change, affecting some of the PRINCIPAL:2 -> PRINCIPAL:5 isoforms.
Our fault, we asked for the change!
We merged and compared @ensembl.org / @gencodegenes.bsky.social , RefSeq and UniProtKB coding genes and investigated the agreements and discrepancies.
Details of what we found in the thread ...

We merged and compared @ensembl.org / @gencodegenes.bsky.social , RefSeq and UniProtKB coding genes and investigated the agreements and discrepancies.
Details of what we found in the thread ...
It's not just budgets but research, institutions, expertise, and training the next generation.
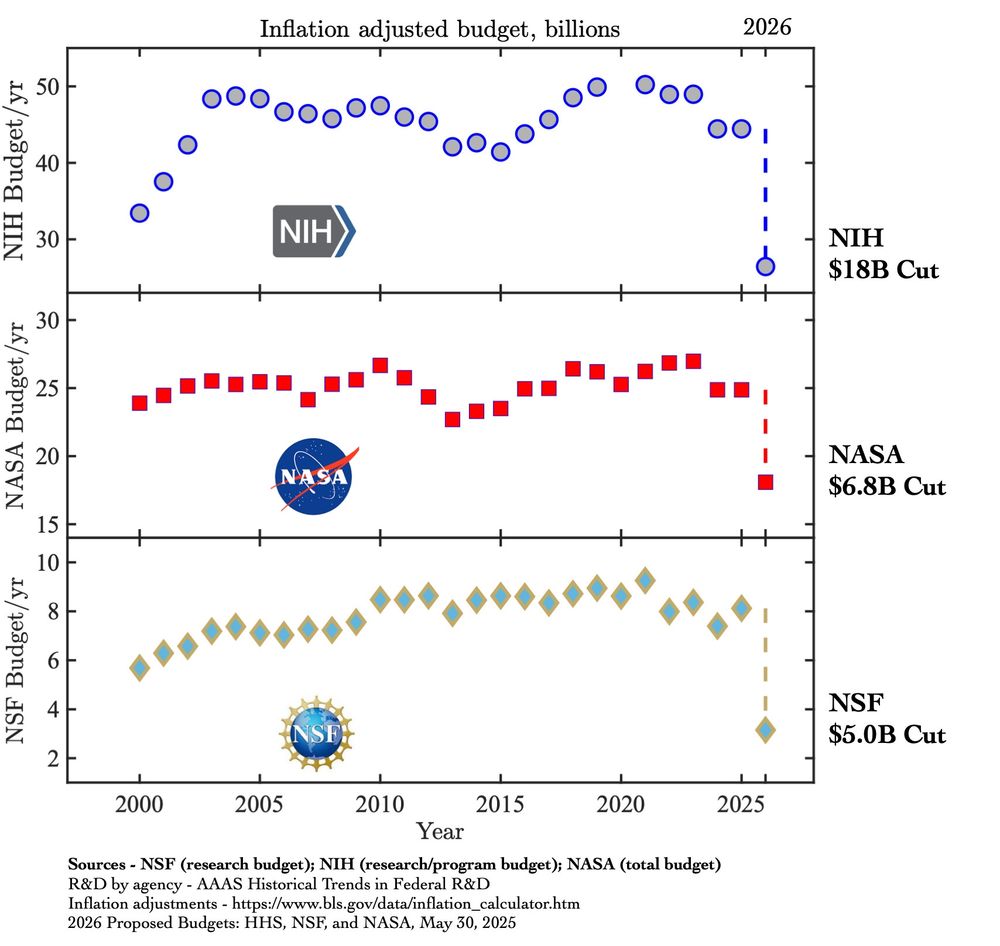
It's not just budgets but research, institutions, expertise, and training the next generation.
“I don’t know how to make this clearer.. If a study has a foreign site, we need to start closing it down or finding a different way to fund it that can be tracked properly,” Matthew Memoli wrote to staff.
“I don’t know how to make this clearer.. If a study has a foreign site, we need to start closing it down or finding a different way to fund it that can be tracked properly,” Matthew Memoli wrote to staff.
Agency staff have also been instructed to not speak about this funding freeze to grantees and applicants
Agency staff have also been instructed to not speak about this funding freeze to grantees and applicants
New this week: NIH has begun terminating projects about misinformation. This comes after the agency sent out a data call on March 26.
One cancelled grant: "Cancer misinformation on social media and its correction"
New this week: NIH has begun terminating projects about misinformation. This comes after the agency sent out a data call on March 26.
One cancelled grant: "Cancer misinformation on social media and its correction"
Starting to see a pattern here ...
www.theguardian.com/us-news/2025...

Starting to see a pattern here ...
www.theguardian.com/us-news/2025...
All of them. Research on pandemic viruses has now ground to a screeching halt in the US.
www.nature.com/articles/d41...
All of them. Research on pandemic viruses has now ground to a screeching halt in the US.
www.nature.com/articles/d41...
www.nytimes.com/2025/03/22/o...

www.nytimes.com/2025/03/22/o...
“As it is, we have some cute-looking hairy mice, and we have learned little biology.”
www.theguardian.com/science/2025...

“As it is, we have some cute-looking hairy mice, and we have learned little biology.”
www.theguardian.com/science/2025...
golikehellmachine.com/doge-is-four...

golikehellmachine.com/doge-is-four...
These principal isoforms are exclusive to the human gene set and are principal isoforms that have been manually corrected. In GENCODE v47 there were 45 genes with Principal:M isoforms.
Here is DCD:

These principal isoforms are exclusive to the human gene set and are principal isoforms that have been manually corrected. In GENCODE v47 there were 45 genes with Principal:M isoforms.
Here is DCD:
in this release 80.7% of human coding genes have a PRINCIPAL:1 isoform.
Also, we have begun the manual curation of principal isoforms. More soon.
@gencodegenes.bsky.social @ensembl.bsky.social
in this release 80.7% of human coding genes have a PRINCIPAL:1 isoform.
Also, we have begun the manual curation of principal isoforms. More soon.
@gencodegenes.bsky.social @ensembl.bsky.social
In particular it shows that what was previously thought to be the human WASH1C gene is actually a pseudogene.
bsky.app/profile/mich...
This goes some way to explaining why so little is known about this vital gene, an integral part of the WASH complex.
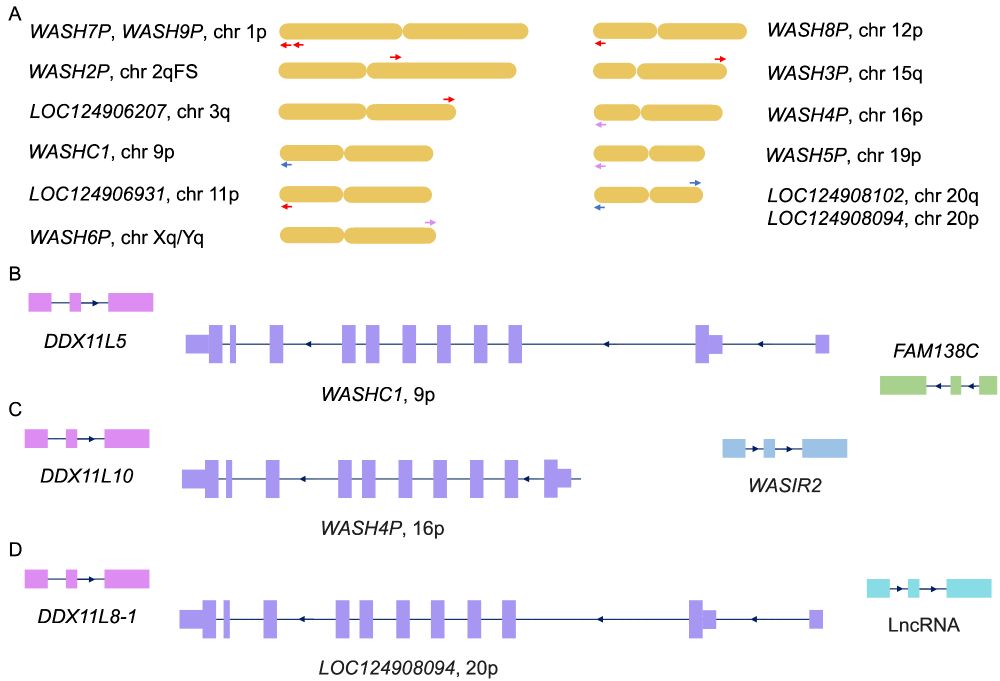
In particular it shows that what was previously thought to be the human WASH1C gene is actually a pseudogene.
bsky.app/profile/mich...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Fascinating.
www.science.org/content/arti...

Fascinating.
www.science.org/content/arti...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
---
#proteomics #prot-preprint

---
#proteomics #prot-preprint


