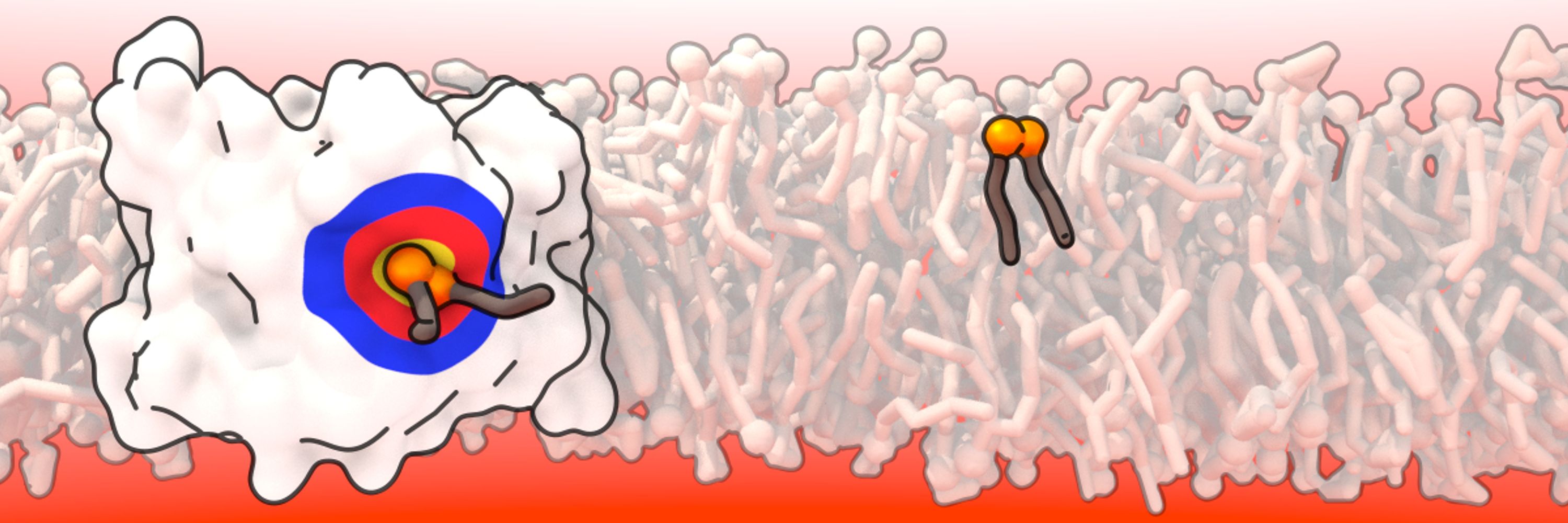
https://www.itqb.unl.pt/labs/multiscale-modeling
pubs.acs.org/doi/10.1021/...
And a big thanks to @tempo3dstudio.bsky.social for the great illustration 🖌️
#compchem #compbiophys #compbiol

pubs.acs.org/doi/10.1021/...
And a big thanks to @tempo3dstudio.bsky.social for the great illustration 🖌️
#compchem #compbiophys #compbiol
See cgmartini.nl for details and how to apply.
Looking forward to seeing you in Groningen, Aug 11-15th.

See cgmartini.nl for details and how to apply.
Looking forward to seeing you in Groningen, Aug 11-15th.
Enjoy!
github.com/MeloLab/Gang...

Enjoy!
github.com/MeloLab/Gang...
(Using the old-but-gold Martini 2 model from @cg-martini.bsky.social)
#compchem #compbiophys #compbiol 🖥️🧪
pubs.acs.org/doi/full/10....
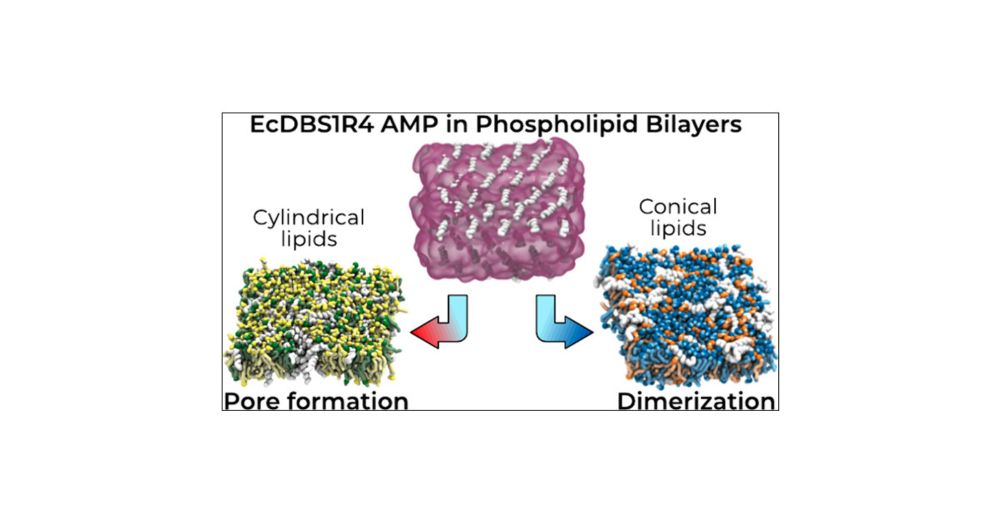
(Using the old-but-gold Martini 2 model from @cg-martini.bsky.social)
#compchem #compbiophys #compbiol 🖥️🧪
pubs.acs.org/doi/full/10....
www.nature.com/articles/s42...

www.nature.com/articles/s42...
Big cheers to the Martini 3 Lipid Taskforce crew, especially Kasper and @pauloctsouza.bsky.social!
#compchem #moleculardynamics
👉 Read the full preprint here: chemrxiv.org/engage/chemr...
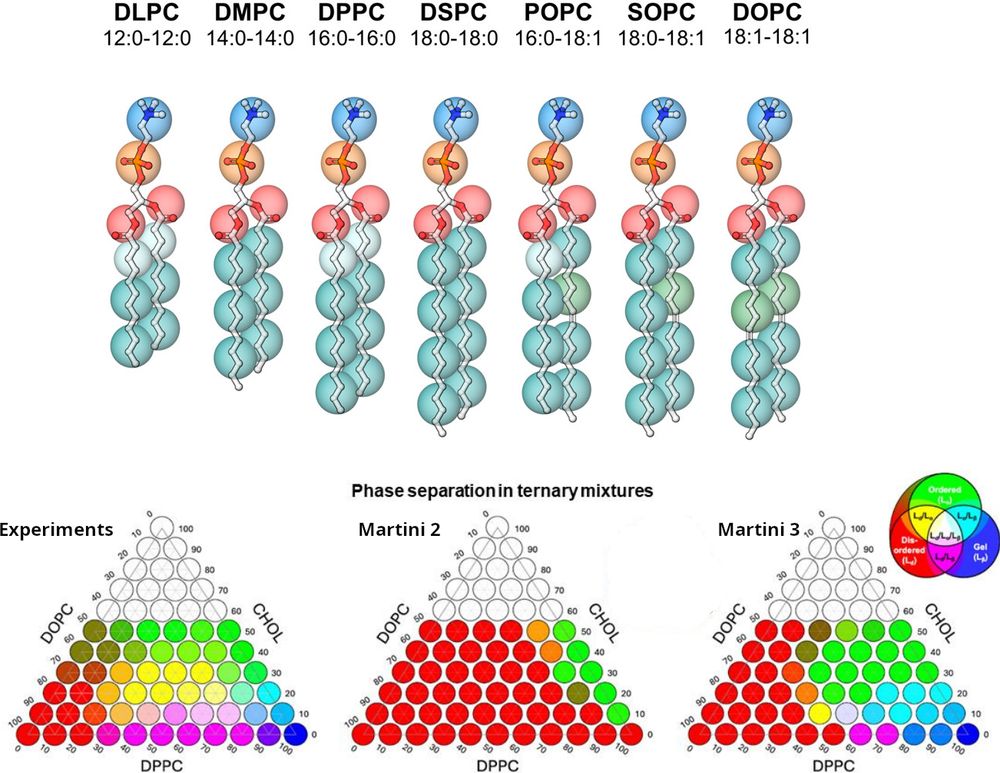
Big cheers to the Martini 3 Lipid Taskforce crew, especially Kasper and @pauloctsouza.bsky.social!
#compchem #moleculardynamics
#CompChem
chemrxiv.org/engage/chemr...

#CompChem
chemrxiv.org/engage/chemr...

Thanks scholargoggler.com 💬☁️

Thanks scholargoggler.com 💬☁️
Read the full article here: rupress.org/jcb/article/...

Read the full article here: rupress.org/jcb/article/...
We just made parameters for those and several other molecules public while the preprint is in the works!
Check out the lab's repository at github.com/MeloLab/Cofa... 🖥️🧑🔬

We just made parameters for those and several other molecules public while the preprint is in the works!
Check out the lab's repository at github.com/MeloLab/Cofa... 🖥️🧑🔬
Hope you all join soon!
Hope you all join soon!
Congrats, @SilvaPereiraLab! https://twitter.com/itqbnova/status/1747948949023969598
Congrats, @SilvaPereiraLab! https://twitter.com/itqbnova/status/1747948949023969598
https://pubs.acs.org/doi/full/10.1021/acs.jctc.3c00547
Big shout out to @SouzaPauloCT and all the collaborators behind this huge effort!
https://pubs.acs.org/doi/full/10.1021/acs.jctc.3c00547
Big shout out to @SouzaPauloCT and all the collaborators behind this huge effort!

https://chemrxiv.org/engage/chemrxiv/article-details/64677ce5fb40f6b3eed8bb26
Big cheers, @SouzaPauloCT, @CG_Martini and the many others in this amazing collaborative endeavor!

https://chemrxiv.org/engage/chemrxiv/article-details/64677ce5fb40f6b3eed8bb26
Big cheers, @SouzaPauloCT, @CG_Martini and the many others in this amazing collaborative endeavor!
https://t.co/fD0fS2ujay
Also, since last tweet, the lab has
i) been at the Martini dev meeting in Malta 😍🍸
ii) organized the 2022 @3dBioinfoPT actions 💪
https://t.co/zguVyJQ7MN
Exciting semester!
https://t.co/fD0fS2ujay
Also, since last tweet, the lab has
i) been at the Martini dev meeting in Malta 😍🍸
ii) organized the 2022 @3dBioinfoPT actions 💪
https://t.co/zguVyJQ7MN
Exciting semester!
Check it out 😃
https://www.frontiersin.org/articles/10.3389/fmedt.2022.1009451

Check it out 😃
https://www.frontiersin.org/articles/10.3389/fmedt.2022.1009451



Interested in a PhD in sunny Lisbon with top players in the field? Then check out our suberin modeling project, with a dash of experimental validation, co-hosted with the @SilvaPereiraLab!
https://t.co/NdQcbHtfVw
Interested in a PhD in sunny Lisbon with top players in the field? Then check out our suberin modeling project, with a dash of experimental validation, co-hosted with the @SilvaPereiraLab!
https://t.co/NdQcbHtfVw
And the @MeloLab will be lecturing, to cater to all your Martini coarse-graining needs! https://twitter.com/BioDataPT/status/1450817222247698434
And the @MeloLab will be lecturing, to cater to all your Martini coarse-graining needs! https://twitter.com/BioDataPT/status/1450817222247698434
https://t.co/uygbmTznih
Be sure to check the goods at our development GitHub repo: https://t.co/hPWaxE7Cgp

https://t.co/uygbmTznih
Be sure to check the goods at our development GitHub repo: https://t.co/hPWaxE7Cgp

