
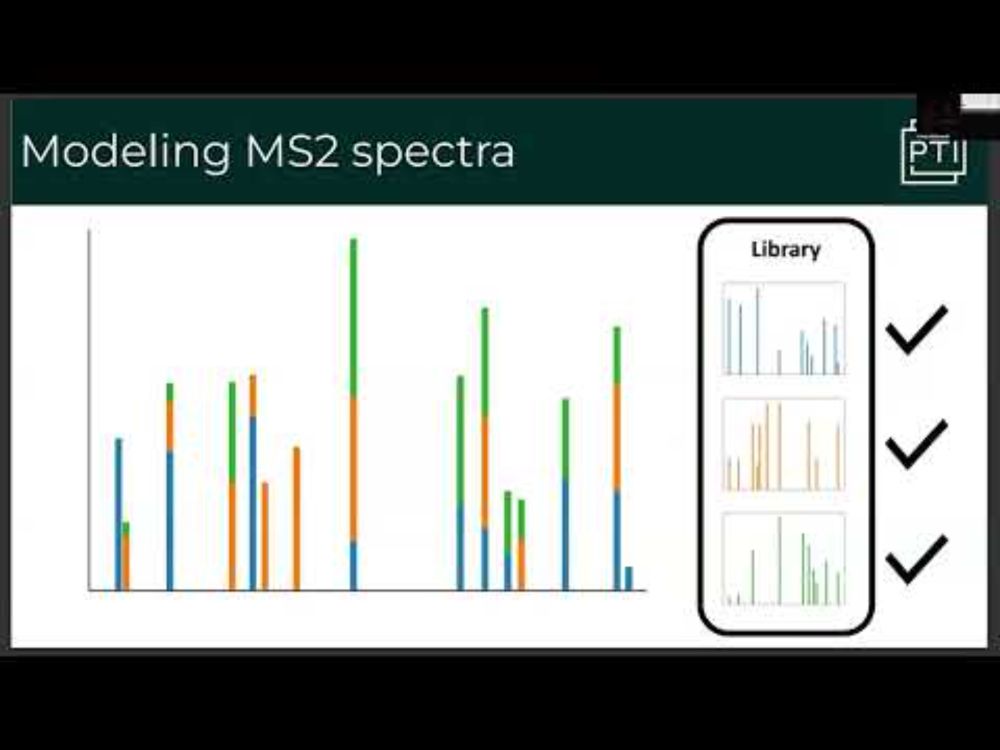
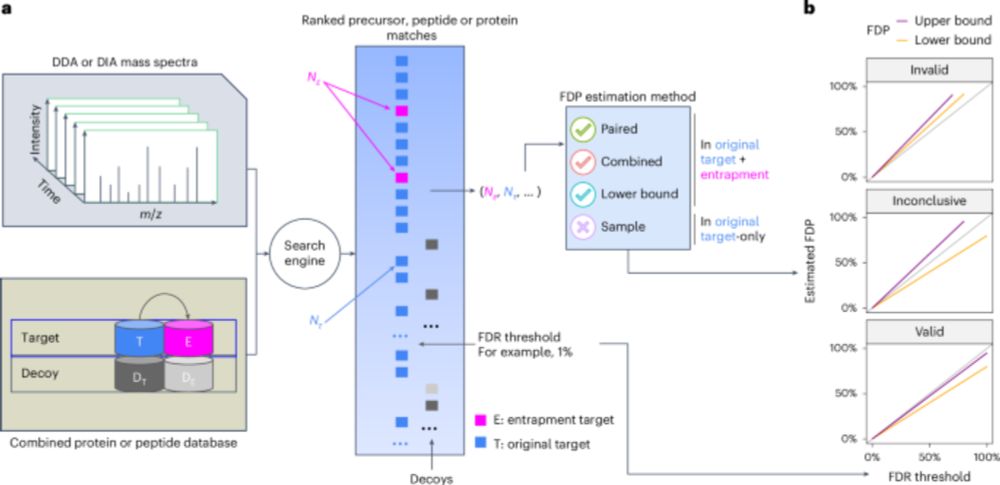
JMod: Joint modeling of mass spectra for enhanced multiplexing in the mass and time domains
youtu.be/qgqloCJJa1M?...
www.nature.com/articles/s41...

🔗 www.msaid.de/conferences/...
#TeamMassSpec

🔗 www.msaid.de/conferences/...
#TeamMassSpec
#ISO27001 #TÜVSüd #DataSecurity #AI

@msaid-de.bsky.social @kusterlab.bsky.social www.nature.com/articles/s41...

@msaid-de.bsky.social @kusterlab.bsky.social www.nature.com/articles/s41...
#CHIMERYS has already transformed #proteomics since 2022, powering DDA, DIA & PRM analysis in a single workflow.
Wanna know how CHIMERYS works? 👉 www.nature.com/articles/s41...
#TeamMassSpec #NatureMethods #ASMS2025 🧪🔬
1/7

📅 April 1 | 🕒 17:00
🔗 Register here: events.teams.microsoft.com/event/26ff8b...

📅 April 1 | 🕒 17:00
🔗 Register here: events.teams.microsoft.com/event/26ff8b...


