Phyloformer 2, a deep end-to-end phylogenetic reconstruction method: arxiv.org/abs/2510.12976
Using neural posterior estimation, it outperforms Phyloformer 1 and maximum-likelihood methods under simple and complex evolutionary models.
🧵1/17
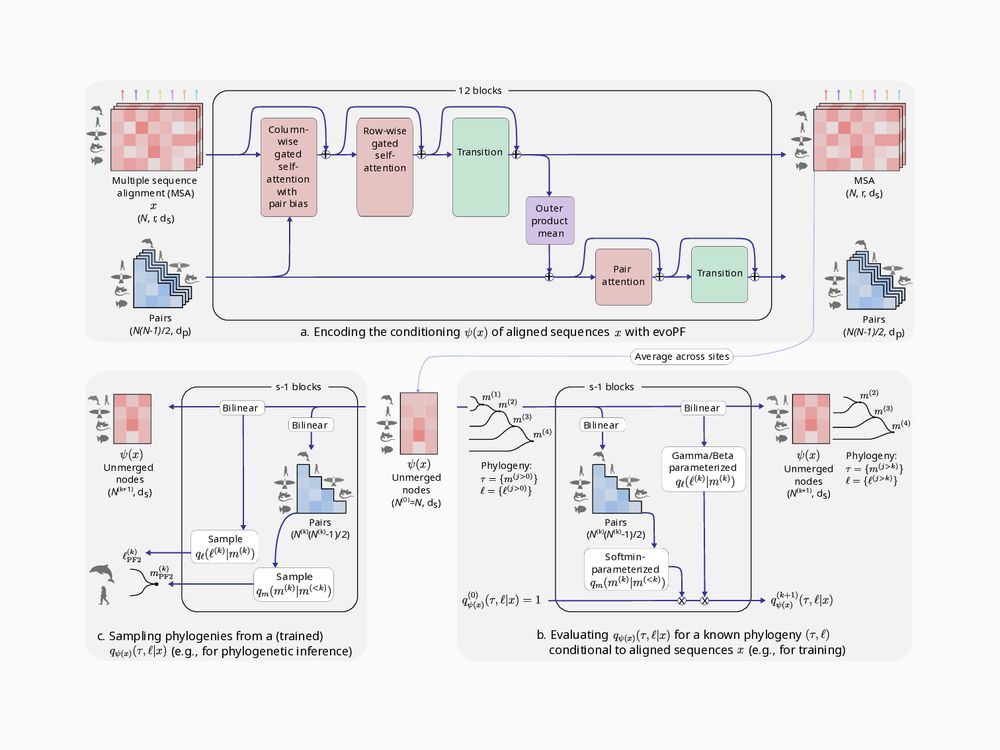
Phyloformer 2, a deep end-to-end phylogenetic reconstruction method: arxiv.org/abs/2510.12976
Using neural posterior estimation, it outperforms Phyloformer 1 and maximum-likelihood methods under simple and complex evolutionary models.
🧵1/17
👉 Preprint www.biorxiv.org/content/10.1...
👉 Github github.com/rolandfaure/...

👉 Preprint www.biorxiv.org/content/10.1...
👉 Github github.com/rolandfaure/...
D’où viennent les fleurs ? lejournal.cnrs.fr/videos/dou-v...

D’où viennent les fleurs ? lejournal.cnrs.fr/videos/dou-v...
@lblassel.bsky.social , assisted by P. Veber, Bastien Boussau
and myself.
The code and data are available at github.com/lucanest/Phy...
Please share if you find this interesting, and we welcome your feedback :)
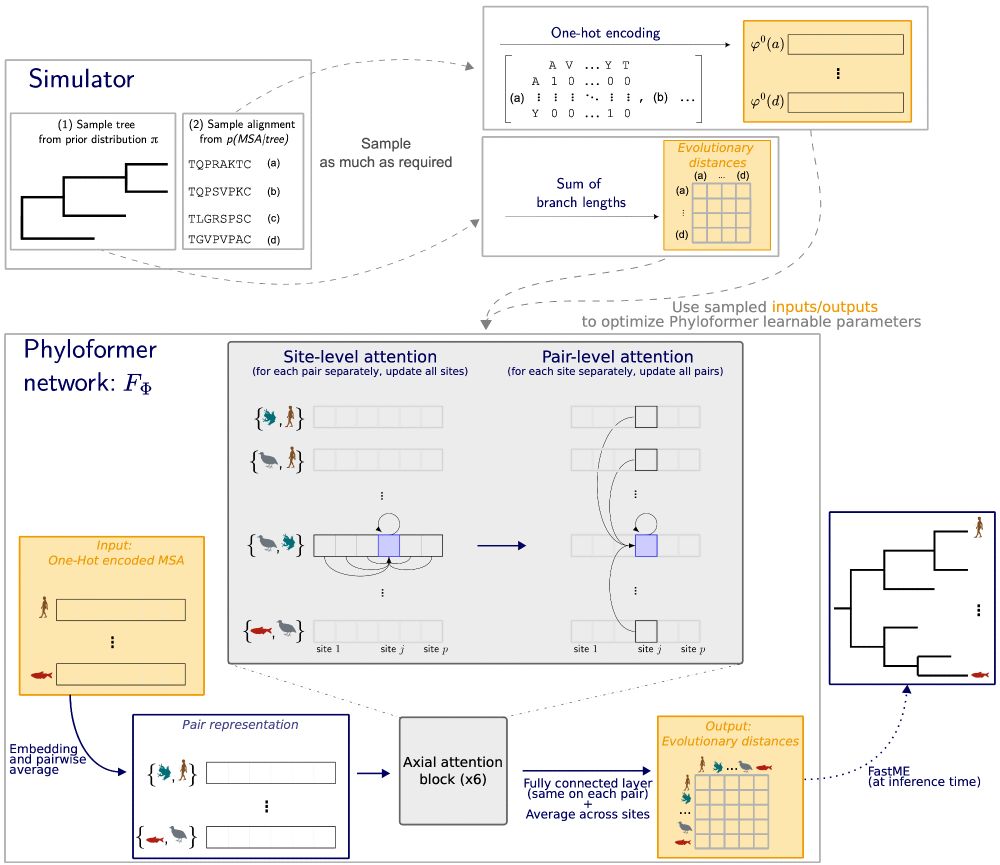
@lblassel.bsky.social , assisted by P. Veber, Bastien Boussau
and myself.
The code and data are available at github.com/lucanest/Phy...
Please share if you find this interesting, and we welcome your feedback :)
Faster than distance methods like neighbor joining, it outperforms maximum likelihood methods under complex models of sequence evolution.
🧵

Faster than distance methods like neighbor joining, it outperforms maximum likelihood methods under complex models of sequence evolution.
🧵

