
https://go.nature.com/4jGqiUc

https://go.nature.com/4jGqiUc

When NIH cancels "diversity" grants (e.g., diversity supplements, Fs, Ks, etc), they aren't canceling the grants based on *what* the grant studies.
They're cancelling them based on *who* is doing the research. And that disallowed "who" is anyone not white.
When NIH cancels "diversity" grants (e.g., diversity supplements, Fs, Ks, etc), they aren't canceling the grants based on *what* the grant studies.
They're cancelling them based on *who* is doing the research. And that disallowed "who" is anyone not white.
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Bret Payseur
academic.oup.com/jeb/advance-...
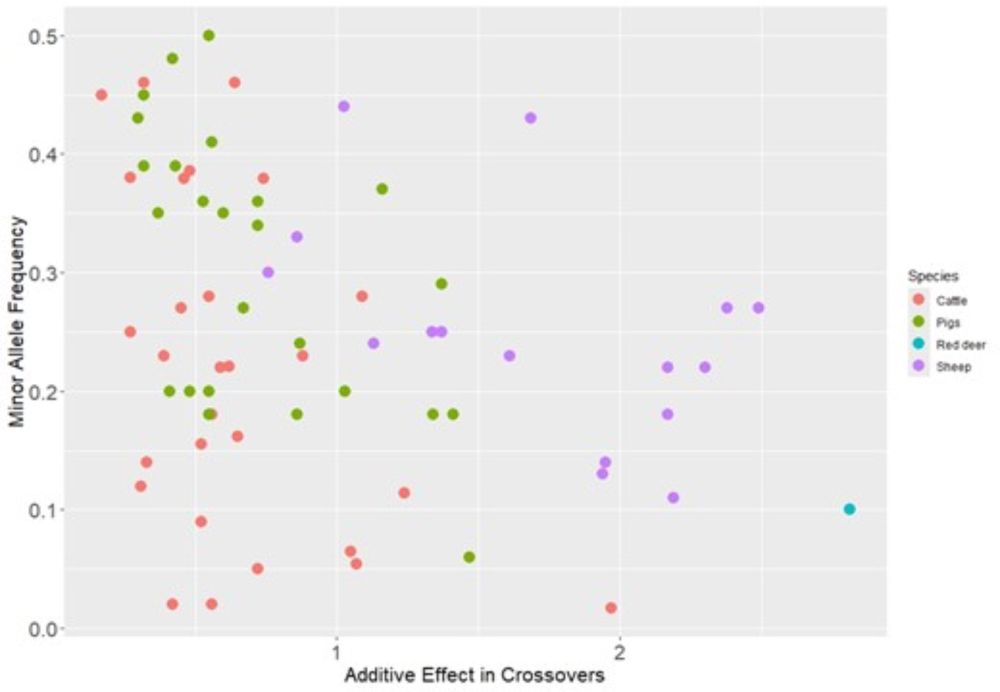
Bret Payseur
academic.oup.com/jeb/advance-...
Selection Can Favor a Recombination Landscape That Limits Polygenic Adaptation
academic.oup.com/mbe/article/...
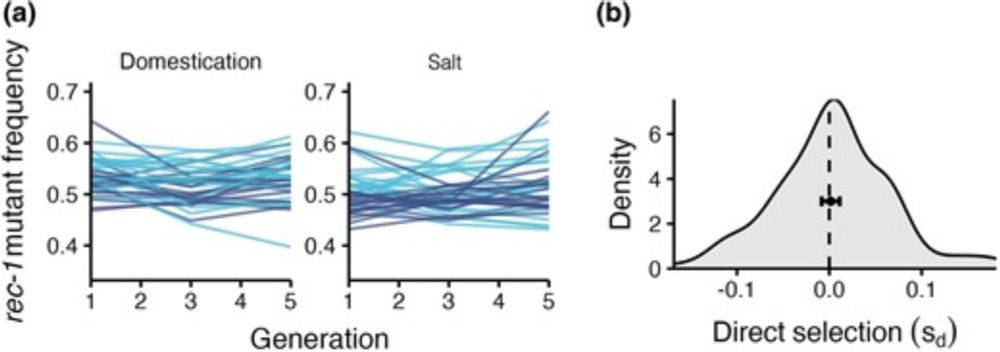
Selection Can Favor a Recombination Landscape That Limits Polygenic Adaptation
academic.oup.com/mbe/article/...
@susanjohnston.bsky.social
and I will be hosting a symposium at
@evolmtg.bsky.social
#evol2024: "Recombination Rate Variation: Causes, Consequences, and Evolution". All are free to apply, see evolutionmeetings.org for details! Please share/RT.

@susanjohnston.bsky.social
and I will be hosting a symposium at
@evolmtg.bsky.social
#evol2024: "Recombination Rate Variation: Causes, Consequences, and Evolution". All are free to apply, see evolutionmeetings.org for details! Please share/RT.
GATK will now encode missing genotypes as "0/0", or homozygous ref, instead of "./." in VCF files. To identify sites with missing data, you'll have to check the DP field.

GATK will now encode missing genotypes as "0/0", or homozygous ref, instead of "./." in VCF files. To identify sites with missing data, you'll have to check the DP field.
Details can be found here: aprecruit.ucr.edu/JPF01846
I am happy to answer questions (I'm on the search committee, but not chair)
Details can be found here: aprecruit.ucr.edu/JPF01846
I am happy to answer questions (I'm on the search committee, but not chair)


