Kops Lab
@kopslab.bsky.social
120 followers
85 following
16 posts
Interested in all things chromosome segregation and aneuploidy | Director & PI at Hubrecht Institute
Posts
Media
Videos
Starter Packs
Reposted by Kops Lab
Kops Lab
@kopslab.bsky.social
· Jul 9
Kops Lab
@kopslab.bsky.social
· Jul 9
Kops Lab
@kopslab.bsky.social
· Jul 9
Kops Lab
@kopslab.bsky.social
· Jul 9
Kops Lab
@kopslab.bsky.social
· Jul 9
Kops Lab
@kopslab.bsky.social
· Jul 9
Kops Lab
@kopslab.bsky.social
· Jul 9
Kops Lab
@kopslab.bsky.social
· Jul 9
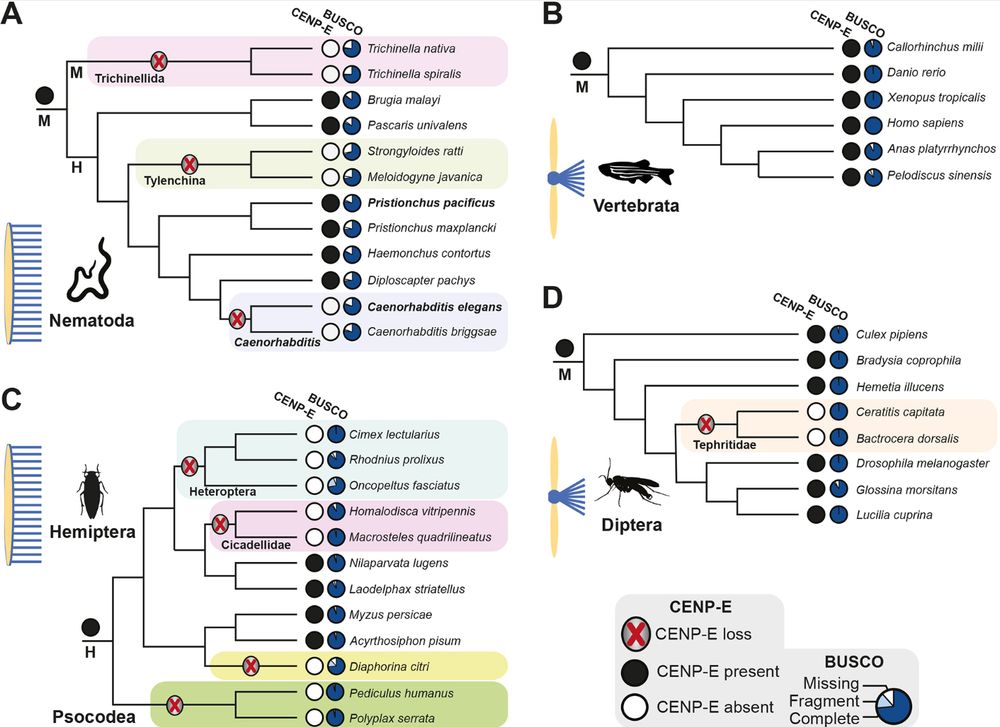
Cancer type-specific association of p53 deficiency with aneuploidy and chromosome losses
Aneuploidy and mutations in the TP53 tumor suppressor gene are among the most frequent genetic alterations in cancer, and p53 inactivation is considered an important contributor to the emergence of cancer aneuploidy. It is unclear, however, if p53 protects against particular forms of copy number alterations and whether it does so universally across cancer types. By analyzing p53 status and various aneuploidy features in 31 cancer types in the TCGA database, we verify that on a pan-cancer level p53-deficient cancers tend to have a higher degree of aneuploidy. However, for many cancer types, the average degree of aneuploidy is similar in p53-proficient and -deficient samples, and a substantial degree of aneuploidy can accumulate with intact p53 in almost all cancer types. Neither arm-level nor whole chromosome aneuploidy but rather chromosome loss events distinguish p53-deficient from proficient cancers. p53 inactivation is therefore neither sufficient nor necessary for the emergence of cancer aneuploidy, but is associated with the degree of aneuploidy in a subset of cancer types and more universally with chromosome losses. Our findings underscore the poorly understood nature of aneuploidy emergence in cancer and shed new light on the role of p53 therein. ### Competing Interest Statement The authors have declared no competing interest. Dutch Cancer Society (KWF Kankerbestrijding) European Research Council, ERC-SyG 855158
www.biorxiv.org
Kops Lab
@kopslab.bsky.social
· Mar 25
We closed today with a great keynote by @embo.org director Fiona Watt, highlighting how spatial maps of epidermal cell populations have allowed the generation and testing of new hypotheses.
Thanks for joining the #HubrechtSymposium2025! We hope you leave with fresh insights and new connections.
Thanks for joining the #HubrechtSymposium2025! We hope you leave with fresh insights and new connections.



Reposted by Kops Lab
Reposted by Kops Lab
Reposted by Kops Lab