Kalki Kukreja
@kkukreja.bsky.social
480 followers
210 following
12 posts
Stem cell and devbio scientist
Posts
Media
Videos
Starter Packs
Reposted by Kalki Kukreja
Reposted by Kalki Kukreja
Reposted by Kalki Kukreja
Reposted by Kalki Kukreja
Reposted by Kalki Kukreja
Reposted by Kalki Kukreja
Kalki Kukreja
@kkukreja.bsky.social
· Dec 19
James Briscoe
@jamesbriscoe.bsky.social
· Dec 19
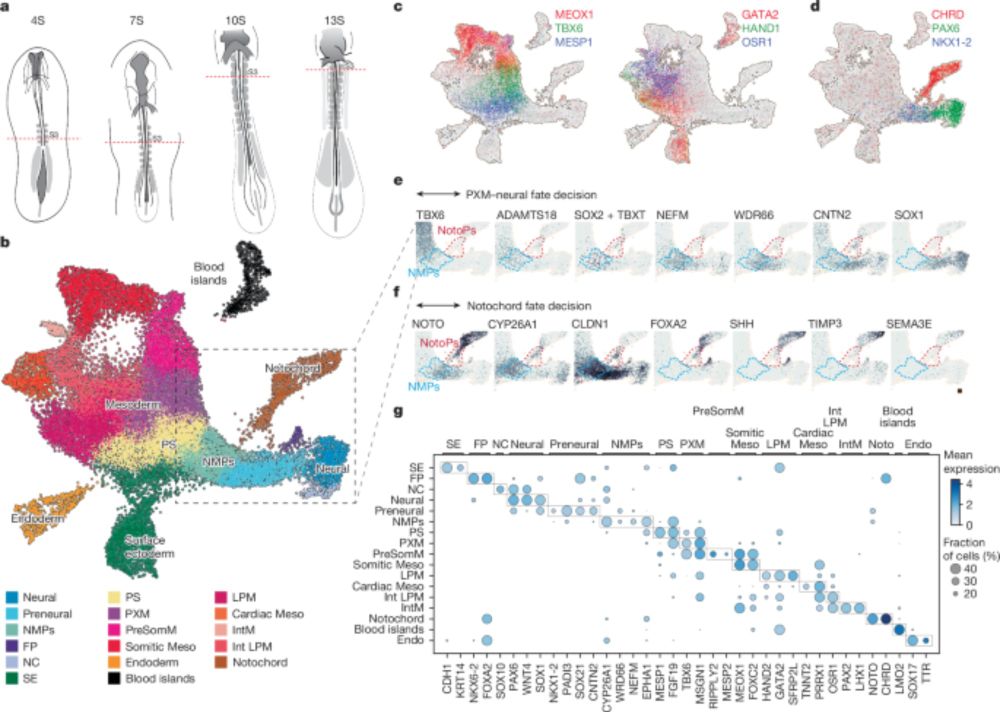
Timely TGFβ signalling inhibition induces notochord - Nature
Through analysis of developing chick embryos and in vitro differentiation of embryonic stem cells, a study develops a method to generate a model of the human trunk with a notochord.
www.nature.com
Kalki Kukreja
@kkukreja.bsky.social
· Dec 12
Reposted by Kalki Kukreja
Kalki Kukreja
@kkukreja.bsky.social
· Nov 28
Kalki Kukreja
@kkukreja.bsky.social
· Nov 25
Reposted by Kalki Kukreja
陳昱嘉 Yu-Chia Chen
@evodevo-tw.bsky.social
· Nov 23
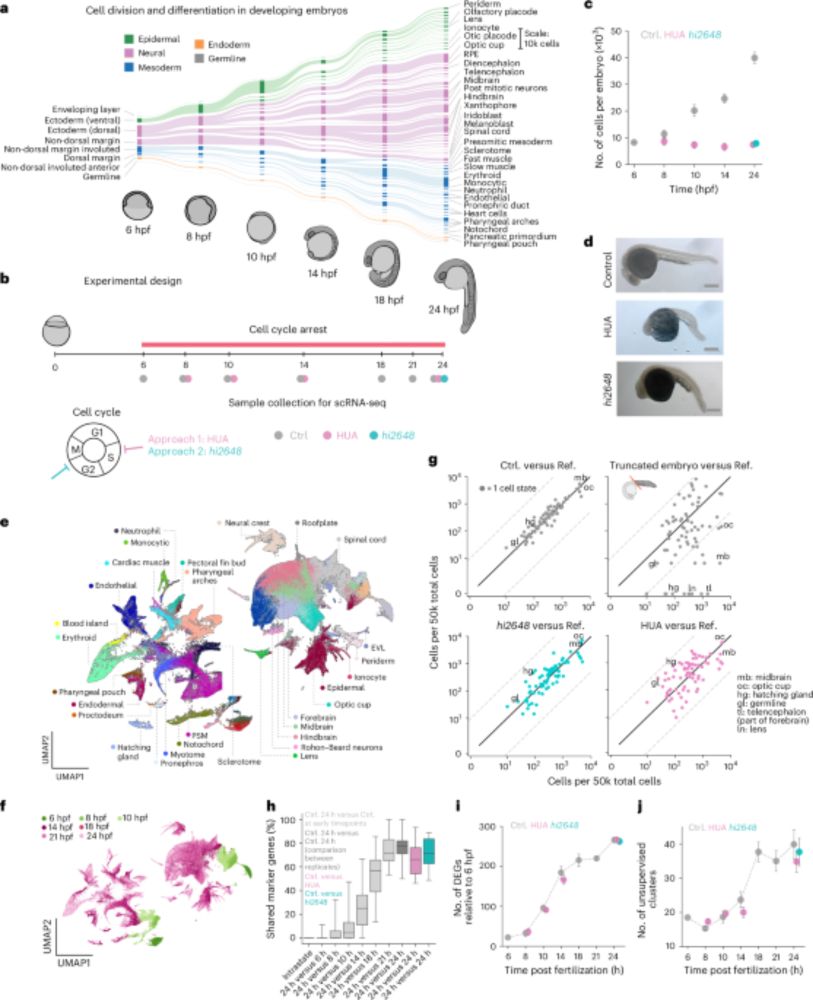
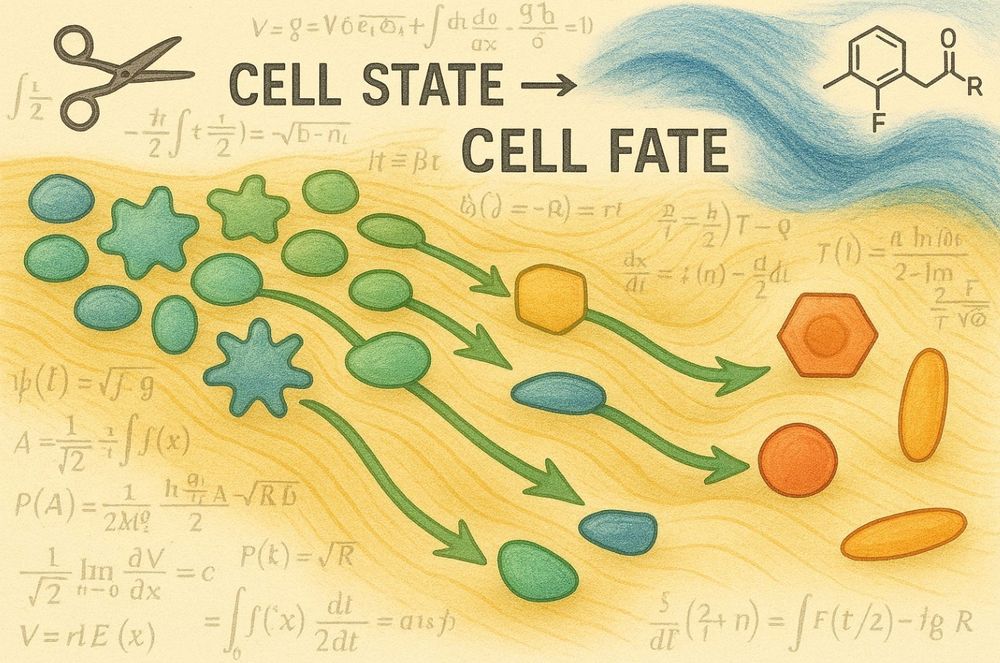
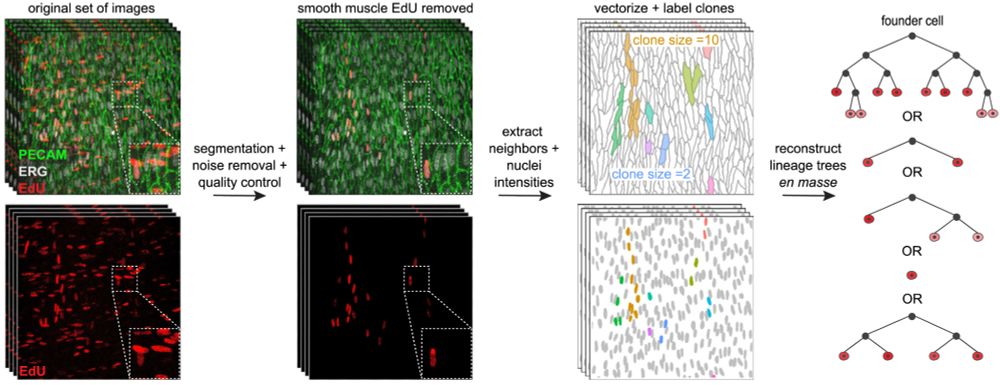
Cell state transitions are decoupled from cell division during early embryo development - Nature Cell Biology
Kukreja et al. show that blocking cell division in zebrafish does not affect differentiation of major cell types during gastrulation and segmentation, but it does decelerate differentiation of particu...
www.nature.com
Kalki Kukreja
@kkukreja.bsky.social
· Nov 18
Kalki Kukreja
@kkukreja.bsky.social
· Nov 18
Kalki Kukreja
@kkukreja.bsky.social
· Nov 18