https://academic.oup.com/jxb
[email protected]

🌾 On the cover: Heterosis has been shown to increase seed yield, produce bigger seeds, reduce germination time and make seeds more tolerant/resistant. See Belcapo et al. 📝
🔗👉 Read the full issue now: academic.oup.com/jxb...
#PlantScience 🧪
🌱 "Adaptation of seeds to climate change is promoted by the mother plant". 🌱
Steven Penfield highlights this research recently published in JXB by Fernández Farnocchia et al.
Insight 🔗 doi.org/10.1093/jxb/...
Research 🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

🌱 "Adaptation of seeds to climate change is promoted by the mother plant". 🌱
Steven Penfield highlights this research recently published in JXB by Fernández Farnocchia et al.
Insight 🔗 doi.org/10.1093/jxb/...
Research 🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
In this technical advance, Decouard et al. show how to identify plant metabolites in root exudates specifically produced in the presence of arbuscular mycorrhizal fungi in an aeroponic-based system.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
![Fig. 1.Description of the aeroponics cultivation system and downstream collection and analysis of root exudates. (A) Scheme of the experimental setup [the germinated spores of the AMF Rhizophagus irregularis were created in BioRender [Bouhedi, B. (2025) https://BioRender.com/qtva7lu]. (B) Pictures of aerial (upper panel) and root parts (lower panel) of maize plants cultivated in aeroponics at the V4 stage. (C) Workflow of the molecular and cellular analyses performed on aeroponics-grown maize plants and their root exudates. (D) Biological significance and composition of the different samples analyzed by untargeted metabolomics. IRM, intraradical mycelium; ERM, extraradical mycelium; GR24, synthetic analog of strigolactone.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:ggz22gcwkakll7efac23g33s/bafkreig3lyjohudeapfmfqacxpc5uwghvpdzym5hqh2emwqi7fotn2pi2a@jpeg)
In this technical advance, Decouard et al. show how to identify plant metabolites in root exudates specifically produced in the presence of arbuscular mycorrhizal fungi in an aeroponic-based system.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
López-Gómez et al. review the role of protein–protein interactions in flower and fruit development and discuss the latest techniques to study them.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪 @defolter-lab.bsky.social

López-Gómez et al. review the role of protein–protein interactions in flower and fruit development and discuss the latest techniques to study them.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪 @defolter-lab.bsky.social
🌏 🌿 Lipid Function in the Plant Environment Special Issue closes on December 31st! 🌿 🌏
Guest editors: Kenji Matsui, Juliette Jouhet, Koichi Kobayashi & Yuki Nakamura 🌿
Got a manuscript? 👉 Contact the JXB office: bit.ly/JXBissues
#JXBspecialissues #PlantScience 🧪


🌏 🌿 Lipid Function in the Plant Environment Special Issue closes on December 31st! 🌿 🌏
Guest editors: Kenji Matsui, Juliette Jouhet, Koichi Kobayashi & Yuki Nakamura 🌿
Got a manuscript? 👉 Contact the JXB office: bit.ly/JXBissues
#JXBspecialissues #PlantScience 🧪
🌾 On the cover: Heterosis has been shown to increase seed yield, produce bigger seeds, reduce germination time and make seeds more tolerant/resistant. See Belcapo et al. 📝
🔗👉 Read the full issue now: academic.oup.com/jxb...
#PlantScience 🧪

🌾 On the cover: Heterosis has been shown to increase seed yield, produce bigger seeds, reduce germination time and make seeds more tolerant/resistant. See Belcapo et al. 📝
🔗👉 Read the full issue now: academic.oup.com/jxb...
#PlantScience 🧪
🌾 On the cover: Heterosis has been shown to increase seed yield, produce bigger seeds, reduce germination time and make seeds more tolerant/resistant. See Belcapo et al. 📝
🔗👉 Read the full issue now: academic.oup.com/jxb...
#PlantScience 🧪

Climate change increasingly exposes plants to stresses that occur simultaneously. In this review, Li et al. examine existing use and the potential role of reflectance spectroscopy in understanding plant responses to combined stresses.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

Climate change increasingly exposes plants to stresses that occur simultaneously. In this review, Li et al. examine existing use and the potential role of reflectance spectroscopy in understanding plant responses to combined stresses.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
In this review, Zhou & Dobritsa summarise recent insights into the mechanisms underlying plasma membrane domain formation and pollen aperture development in Arabidopsis and rice.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

In this review, Zhou & Dobritsa summarise recent insights into the mechanisms underlying plasma membrane domain formation and pollen aperture development in Arabidopsis and rice.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
Gupta et al. provide updated insights into how nitrate reductase-mediated nitric oxide production contributes to stress adaptation, with a focus on its regulation, signaling interactions, and potential for enhancing crop resilience.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

Gupta et al. provide updated insights into how nitrate reductase-mediated nitric oxide production contributes to stress adaptation, with a focus on its regulation, signaling interactions, and potential for enhancing crop resilience.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
⚠️🧫🪴 Invisible contaminants, irreversible consequences? LDPE residues twist the Arabidopsis holobiome. Chakraborty & Guo comment on research recently published in JXB by Lee et al.
Insight 🔗 doi.org/10.1093/jxb/...
Research 🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
![Fig. 1 (shortened, full legend in paper): Invisible stress: molecular disruptions beneath the surface. Microplastic contamination from LDPE residues in soil presents a deceptive paradox (A): plants may appear healthy (B), yet their molecular machinery tells another story (C). The study by Lee et al. (2025) reveals a striking transcriptome–phenotype disconnect in Arabidopsis thaliana. Despite maintaining biomass and morphology, exposed plants exhibited widespread suppression of photosynthesis-related genes [e.g. chlorophyll a/b-binding protein (CAB), small subunit of Rubisco (rbcS)] and nutrient transporter genes (NRT2.5), alongside shifts in the rhizosphere microbial community toward plastic-degrading taxa (C). These findings suggest that traditional indicators such as yield or chlorophyll content may be insufficient to detect early-stage plastic-induced stress (D).](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:ggz22gcwkakll7efac23g33s/bafkreibj4e5j7xbm6ibvlnt3ss2x2jcnwm2hqtbma7oxzer646iigchg24@jpeg)
⚠️🧫🪴 Invisible contaminants, irreversible consequences? LDPE residues twist the Arabidopsis holobiome. Chakraborty & Guo comment on research recently published in JXB by Lee et al.
Insight 🔗 doi.org/10.1093/jxb/...
Research 🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

🌾 On the cover: Heterosis has been shown to increase seed yield, produce bigger seeds, reduce germination time and make seeds more tolerant/resistant. See Belcapo et al. 📝
🔗👉 Read the full issue now: academic.oup.com/jxb...
#PlantScience 🧪

🌾 On the cover: Heterosis has been shown to increase seed yield, produce bigger seeds, reduce germination time and make seeds more tolerant/resistant. See Belcapo et al. 📝
🔗👉 Read the full issue now: academic.oup.com/jxb...
#PlantScience 🧪
I've collected recent advances in transcriptional and epigenetic 🧬 Regulation of Chloroplast Biogenesis and Differentiation url: academic.oup.com/jxb/article/...

I've collected recent advances in transcriptional and epigenetic 🧬 Regulation of Chloroplast Biogenesis and Differentiation url: academic.oup.com/jxb/article/...
The tomato dad1 mutant lacks jasmonate biosynthesis, forming seedless fruits. Expression data shows spatiotemporal localization of jasmonate regulates fruit set primarily by modulating gibberellin metabolism - Nomura et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

The tomato dad1 mutant lacks jasmonate biosynthesis, forming seedless fruits. Expression data shows spatiotemporal localization of jasmonate regulates fruit set primarily by modulating gibberellin metabolism - Nomura et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
🍅 Genome-edited tomato mutants for the SPATULA gene elucidated conserved functions during gynoecium development, improving our understanding of the SPT gene in fleshy fruits- Martínez-Estrada et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪 @defolter-lab.bsky.social
![Fig. 4.Effect of the lack of SlSPT function on pollen tube growth and ovary characteristics in slspt mutants and wild-type (WT; Micro-Tom) plants. (A) Pollen tube growth in the gynoecium. (B) Pollen tube staining measured as relative intensity units. (C) Micrographs of transverse sections of the ovary at anthesis, with a magnified view of the pericarp of the ovary at anthesis. (D, E) Comparison of cell layers and pericarp thickness between WT and slspt mutants. Different letters indicate statistically significant differences [ANOVA followed by Fisher's LSD test, P≤0.05, n=10, for (B), Kruskal–Wallis test followed by Dunn’s test, P≤0.05, n=15, for (D); Tukey’s range test, P≤0.05, n=15, for (E)]. Scale bars: 1 mm (A), 0.4 mm (C left); 50 µm (C right).](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:ggz22gcwkakll7efac23g33s/bafkreiaf3ebstlmcdbo5jov4d2slkfczlrqqb4wtsci55jroaspvt7nk4i@jpeg)
🍅 Genome-edited tomato mutants for the SPATULA gene elucidated conserved functions during gynoecium development, improving our understanding of the SPT gene in fleshy fruits- Martínez-Estrada et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪 @defolter-lab.bsky.social
The nutritional quality of fresh fruit is not just a matter of bioactive content but of all the structural and biochemical properties that develop from the field to the digestive tract - Bertin et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

The nutritional quality of fresh fruit is not just a matter of bioactive content but of all the structural and biochemical properties that develop from the field to the digestive tract - Bertin et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
Wang & de Maagd discuss review recent progress and address remaining questions in relation to tomato fruit development and ripening transcriptional regulation, and highlight future challenges.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

Wang & de Maagd discuss review recent progress and address remaining questions in relation to tomato fruit development and ripening transcriptional regulation, and highlight future challenges.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
MYB and WRKY transcription factors collaboratively regulate suberin biosynthesis in the tomato root exodermis. Antagonistic interactions may fine-tune suberization or act as a brake on overaccumulation - Jo et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
![Fig. 2 (shortened, full legend in paper): Perturbations of SlMYB41, SlMYB92, and SlWRKY71 expression affect the suberization of tomato exodermis. (A) Percentage of cross-sections showing a non-suberized, an early suberized (suberin in <50% of exodermal cells), or a late suberized (suberin in >50% of exodermal cells) exodermis phenotype in hairy root cultures of the control (empty vector), CRISPR knockout lines [Slmyb41-ko, Slmyb92-ko, Slwrky71-ko(4), and Slwrky71-ko(5)], and overexpressing lines (SlMYB41-OX, SlMYB92-OX, and SlWRKY71-OX). The number above the bars represents the number (n) of individual roots examined in each genotype. Asterisks denotes statistically significant differences between each line in comparison with the empty vector control, whereas n.s. indicates no significant differences (P<0.05, χ2 test).](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:ggz22gcwkakll7efac23g33s/bafkreidbanhpxnb52kuljrltp3voz7jfnkuh4js6tbmdxudjsbfxuozteu@jpeg)
The wild tomato Solanum pennellii has specialized leaf design & photosynthetic machinery that boost photosynthetic capacity, which can be transferred to commercial tomato plants via non-GM grafting methods- Ganie et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

The wild tomato Solanum pennellii has specialized leaf design & photosynthetic machinery that boost photosynthetic capacity, which can be transferred to commercial tomato plants via non-GM grafting methods- Ganie et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
A mechanical treatment applied to tomato seedlings enhances stem width, increases vascular bundle number, promotes early flowering & improves fruit yield, involving auxin & ethylene pathways - Castro-Estrada et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

A mechanical treatment applied to tomato seedlings enhances stem width, increases vascular bundle number, promotes early flowering & improves fruit yield, involving auxin & ethylene pathways - Castro-Estrada et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
Tomato cytoplasmic male sterility caused by incompatibility between nuclear and cytoplasmic genetic information is restored through the action of multiple fertility restorer genes of tomato wild relatives - Iki et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

Tomato cytoplasmic male sterility caused by incompatibility between nuclear and cytoplasmic genetic information is restored through the action of multiple fertility restorer genes of tomato wild relatives - Iki et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
MYB and WRKY transcription factors collaboratively regulate suberin biosynthesis in the tomato root exodermis. Antagonistic interactions may fine-tune suberization or act as a brake on overaccumulation - Jo et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
![Fig. 2 (shortened, full legend in paper): Perturbations of SlMYB41, SlMYB92, and SlWRKY71 expression affect the suberization of tomato exodermis. (A) Percentage of cross-sections showing a non-suberized, an early suberized (suberin in <50% of exodermal cells), or a late suberized (suberin in >50% of exodermal cells) exodermis phenotype in hairy root cultures of the control (empty vector), CRISPR knockout lines [Slmyb41-ko, Slmyb92-ko, Slwrky71-ko(4), and Slwrky71-ko(5)], and overexpressing lines (SlMYB41-OX, SlMYB92-OX, and SlWRKY71-OX). The number above the bars represents the number (n) of individual roots examined in each genotype. Asterisks denotes statistically significant differences between each line in comparison with the empty vector control, whereas n.s. indicates no significant differences (P<0.05, χ2 test).](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:ggz22gcwkakll7efac23g33s/bafkreidbanhpxnb52kuljrltp3voz7jfnkuh4js6tbmdxudjsbfxuozteu@jpeg)
MYB and WRKY transcription factors collaboratively regulate suberin biosynthesis in the tomato root exodermis. Antagonistic interactions may fine-tune suberization or act as a brake on overaccumulation - Jo et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
Glandular trichomes function as ultra-fast biomechanical defense triggers in tomatoes, rupturing upon contact with insects and exposing them to sticky and toxic secretions - Popowski et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪 @jaredpopo.bsky.social

Glandular trichomes function as ultra-fast biomechanical defense triggers in tomatoes, rupturing upon contact with insects and exposing them to sticky and toxic secretions - Popowski et al.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪 @jaredpopo.bsky.social
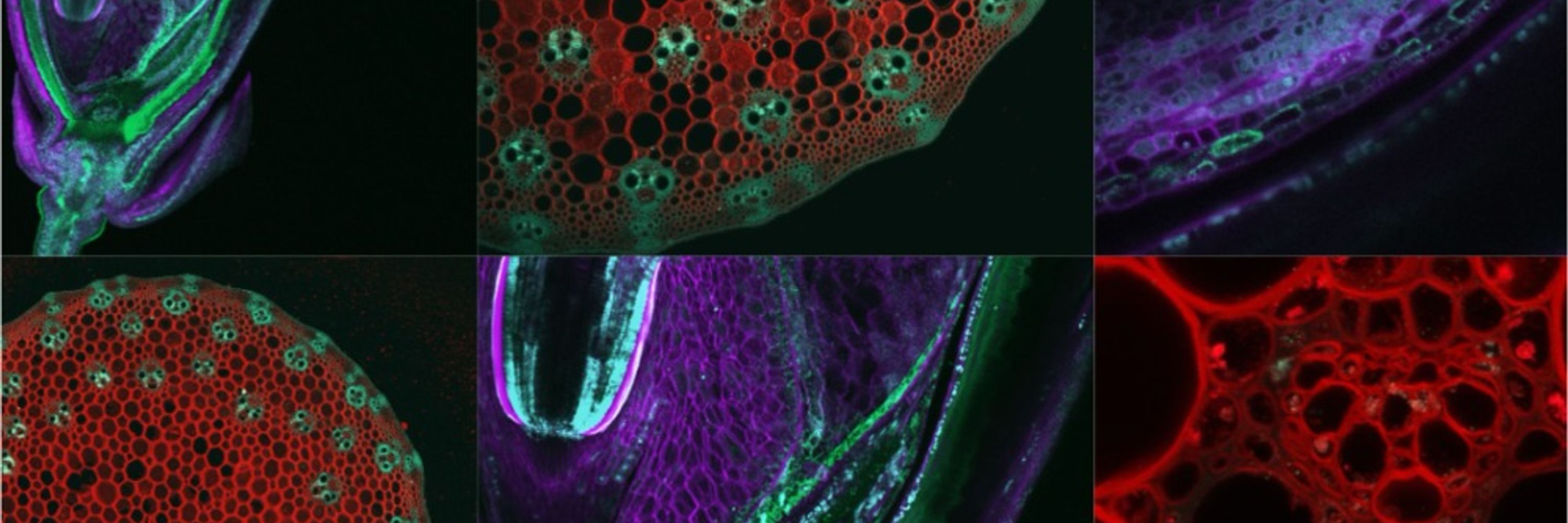
Flores-Tornero et al. have created the first double fluorescent marker line for generative cell and sperm cells of tomato, as well as a protocol to isolate those cells using fluorescence-activated cell sorting 🍅
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

Flores-Tornero et al. have created the first double fluorescent marker line for generative cell and sperm cells of tomato, as well as a protocol to isolate those cells using fluorescence-activated cell sorting 🍅
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪
Heuvelink et al. examine how light intensity, photoperiod, spectrum, directionality and energy regulate whole-plant physiological processes including morphogenesis, source–sink interactions and assimilate partitioning.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

Heuvelink et al. examine how light intensity, photoperiod, spectrum, directionality and energy regulate whole-plant physiological processes including morphogenesis, source–sink interactions and assimilate partitioning.
🔗 doi.org/10.1093/jxb/...
#PlantScience 🧪

