weinstocklab.org

Turns out this 'discovers' classic clonal hematopoiesis drivers and more 🧵👇
www.medrxiv.org/content/10.1...
#ASHG
In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...

In new work @nature.com with @hakha.bsky.social, @jkpritch.bsky.social, and our wonderful coauthors we find that the key factors are what we call Specificity, Length, and Luck!
🧬🧪🧵
www.nature.com/articles/s41...
Turns out this 'discovers' classic clonal hematopoiesis drivers and more 🧵👇
www.medrxiv.org/content/10.1...
#ASHG

Turns out this 'discovers' classic clonal hematopoiesis drivers and more 🧵👇
www.medrxiv.org/content/10.1...
#ASHG
(1) I just opened my lab at Boston Children’s Hospital (Harvard-affiliated)
(2) I’m hiring a postdoc focused on integrating GWAS and functional genomic data. Reach out if you’re interested or connect at ASHG next week!
(3) Learn more at stroberlab.com

(1) I just opened my lab at Boston Children’s Hospital (Harvard-affiliated)
(2) I’m hiring a postdoc focused on integrating GWAS and functional genomic data. Reach out if you’re interested or connect at ASHG next week!
(3) Learn more at stroberlab.com
The E2G Portal! e2g.stanford.edu
This collates our predictions of enhancer-gene regulatory interactions across >1,600 cell types and tissues.
Uses cases 👇
1/
The E2G Portal! e2g.stanford.edu
This collates our predictions of enhancer-gene regulatory interactions across >1,600 cell types and tissues.
Uses cases 👇
1/

www.medrxiv.org/content/10.1...

www.medrxiv.org/content/10.1...
Co-taught with Mike Epstein, Dave Cutler, Karen Conneely, Jingjing Yang, Jian Hu.
Mendelian randomization: weinstocklab.org/lecture_slid...
Biobank scale GWAS methods: weinstocklab.org/lecture_slid...
Hope they're helpful!
Co-taught with Mike Epstein, Dave Cutler, Karen Conneely, Jingjing Yang, Jian Hu.
Mendelian randomization: weinstocklab.org/lecture_slid...
Biobank scale GWAS methods: weinstocklab.org/lecture_slid...
Hope they're helpful!
www.cell.com/cell-genomics/fulltext/S2666-979X(24)00330-6
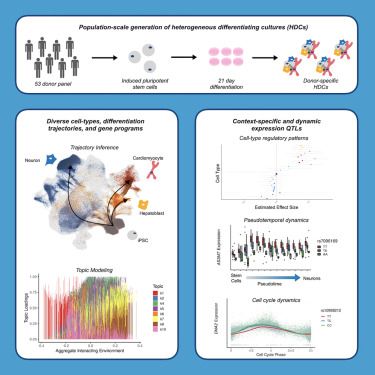
www.cell.com/cell-genomics/fulltext/S2666-979X(24)00330-6
Feel free to DM if of interest.
Feel free to DM if of interest.

