
🔗 academic.oup.com/gbe
🏠 @official-smbe.bsky.social
🤝 @molbioevol.bsky.social
#genome #evolution #science #biology #societyjournal

🔗 doi.org/10.1093/gbe/evaf223
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf210
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf210
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf203
#genome #evolution #migration

🔗 doi.org/10.1093/gbe/evaf203
#genome #evolution #migration
🔗 doi.org/10.1093/gbe/evaf216
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf217
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf217
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf215
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf215
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf211
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf211
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf202
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf202
#genome #evolution
Submit your research and apply for SMBE awards via the submission portal.
📝 Read more & submit here: smbe2026.org/abstracts
And symposium selection is now finalized!
👉 See all accepted symposia: smbe2026.org/programme
#SMBE2026

Submit your research and apply for SMBE awards via the submission portal.
📝 Read more & submit here: smbe2026.org/abstracts
And symposium selection is now finalized!
👉 See all accepted symposia: smbe2026.org/programme
#SMBE2026
🔗 doi.org/10.1093/gbe/evaf216
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf216
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf223
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf223
#genome #evolution
Thanks to @bhuwanabbot.bsky.social
for leading and collaborators:
@raw937.bsky.social @andrabuchan.bsky.social
academic.oup.com/gbe/advance-...

Thanks to @bhuwanabbot.bsky.social
for leading and collaborators:
@raw937.bsky.social @andrabuchan.bsky.social
academic.oup.com/gbe/advance-...
🔗 doi.org/10.1093/gbe/evaf214
#genome #evolution #microbiology

🔗 doi.org/10.1093/gbe/evaf214
#genome #evolution #microbiology
🔗 doi.org/10.1093/gbe/evaf213
#genome #evolution #evodevo

🔗 doi.org/10.1093/gbe/evaf213
#genome #evolution #evodevo
🔗 doi.org/10.1093/gbe/evaf212
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf212
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf200
#genome #evolution #phylogenetics

🔗 doi.org/10.1093/gbe/evaf200
#genome #evolution #phylogenetics
🔗 doi.org/10.1093/gbe/evaf206
#genome #evolution #aDNA #denisova

🔗 doi.org/10.1093/gbe/evaf206
#genome #evolution #aDNA #denisova
🔗 doi.org/10.1093/gbe/evaf207
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf207
#genome #evolution
Read paper in Genome Biology and Evolution: academic.oup.com/gbe/advance-...

Read paper in Genome Biology and Evolution: academic.oup.com/gbe/advance-...
The male-killing gene wmk in Wolbachia splits into 5 types with distinct genomic patterns. Variation in copy number and expression helps explain male-biased lethality across hosts.
Read more: academic.oup.com/gbe/article/...

The male-killing gene wmk in Wolbachia splits into 5 types with distinct genomic patterns. Variation in copy number and expression helps explain male-biased lethality across hosts.
Read more: academic.oup.com/gbe/article/...
🔗 doi.org/10.1093/gbe/evaf208
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf208
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf188
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf188
#genome #evolution
Main findings in the 🧵 below ⬇️
@cpgsthlm.bsky.social @genomebiolevol.bsky.social #aDNA
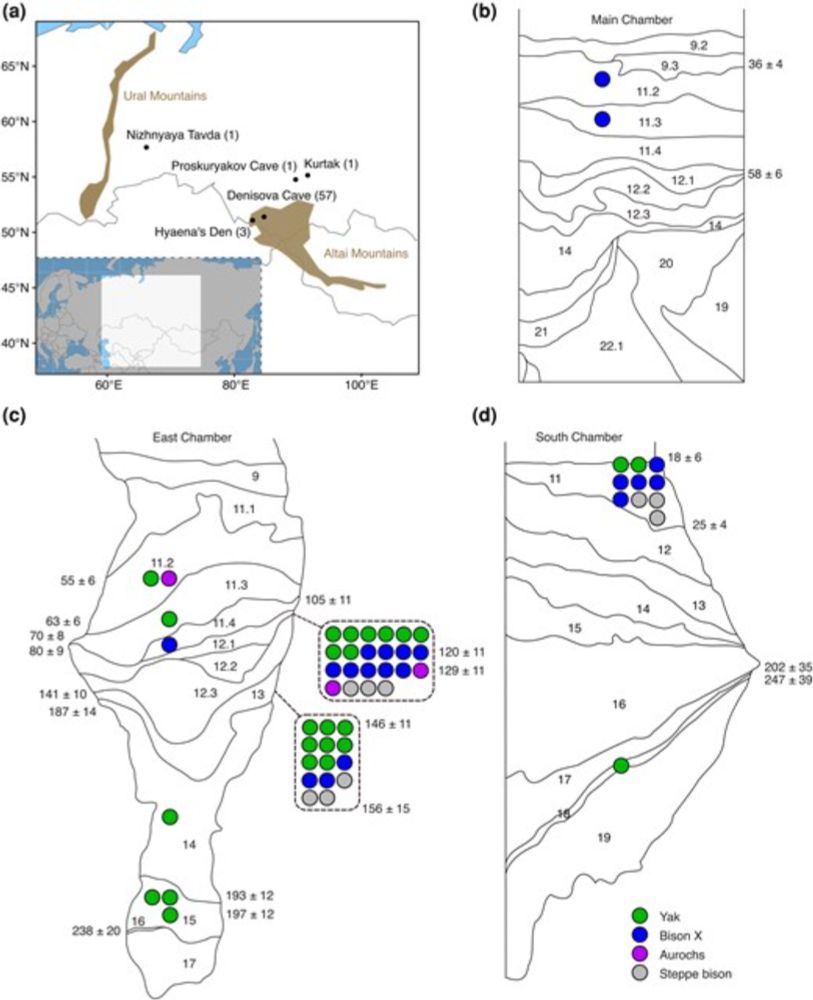
Main findings in the 🧵 below ⬇️
@cpgsthlm.bsky.social @genomebiolevol.bsky.social #aDNA
🔗 doi.org/10.1093/gbe/evaf205
#genome #evolution #birds

🔗 doi.org/10.1093/gbe/evaf205
#genome #evolution #birds
🔗 doi.org/10.1093/gbe/evaf197
#genome #evolution

🔗 doi.org/10.1093/gbe/evaf197
#genome #evolution
🔗 doi.org/10.1093/gbe/evaf199
#genome #evolution #snail


