Fernando Gonzalez-Candelas
@fgonzalef.bsky.social
640 followers
610 following
11 posts
Evolutionary biology, genomic epidemiology, pathogens, phylogenies, phylogenomics, bioinformatics, population genetics and genomics, forensics, and anything to which evolutionary analyses of genes and genomes can be applied to. Squash player (still).
Posts
Media
Videos
Starter Packs
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Rhys White
@rhystwhite.bsky.social
· Feb 13

The need for speed: ultra-rapid high-resolution outbreak analysis in a front-line hospital microbiology laboratory
Many hospital laboratories have technical capacity to perform whole-genome sequencing but lack bioinformatic expertise to analyse sequence data. Sending isolates to reference laboratories creates dela...
doi.org
Reposted by Fernando Gonzalez-Candelas
Wolfgang Huber
@wkhuber.bsky.social
· Jan 24
Reposted by Fernando Gonzalez-Candelas
Matt Jago
@mattjago.bsky.social
· Jan 18
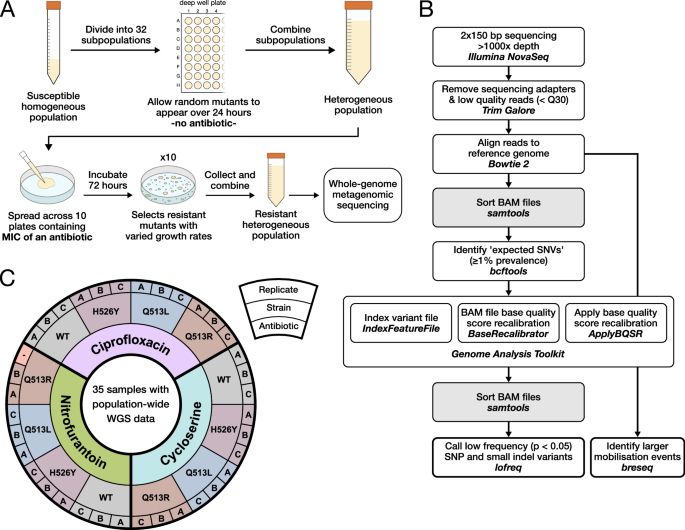
High-throughput method characterizes hundreds of previously unknown antibiotic resistance mutations - Nature Communications
Resistance mutations are challenging to characterize because their effects are highly context dependent. Here, authors present a quantitative mutant screening technique that deconstructs how factors l...
www.nature.com
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Reposted by Fernando Gonzalez-Candelas
Tom Peacock
@peacockflu.bsky.social
· Jan 7

Polymerase mutations underlie adaptation of H5N1 influenza virus to dairy cattle and other mammals.
In early 2024, an unprecedented outbreak of H5N1 high pathogenicity avian influenza was detected in dairy cattle in the USA. The epidemic remains uncontrolled, with spillbacks into poultry, wild birds...
www.biorxiv.org
Reposted by Fernando Gonzalez-Candelas











![screenshot of the new webpage:
[Title] Newly discovered betacoronavirus, Wuhan 2019-2020
A previously unknown betacoronavirus was detected in patients during an outbreak of respiratory illnesses, including atypical pneumonia, that started mid-December 2019 in the city of Wuhan, the capital of Central China's Hubei Province.
The newly discovered coronavirus is similar to some of the betacoronaviruses detected in bats, but it is distinct from SARS-CoV and MERS-CoV.
The genome of the newly discovered CoV consists of a single, positive-stranded RNA that is approximately 30k nucleotides long. The overall genome organization of the newly discovered CoV is similar to that of other coronaviruses. The newly sequenced virus genome encodes the open reading frames (ORFs) common to all betacoronaviruses, including ORF1 ab that encodes many enzymatic proteins, the spike-surface glycoprotein (S), the small envelope protein (E), the matrix protein (M), and the nucleocapsid protein (N), as well as several nonstructural proteins.
[Table]
Virus name
Accession ID
Passage det Collection date
Host
Originating lab
BetaCoV/Wuhan/IVDC-HB-01/2019 EPIISL_402119 Virus Isolate
2019-12-30
Human
National Institute for Viral Disease Control and Prevention, China CDC
BetaCoV/Wuhan/IVDC-HB-04/2020
EPIISL_402120 Original
2020-01-01
Human
National Institute for Viral Disease Control and Prevention, China CDC
BetaCoV/Wuhan/IVDC-HB-05/2019
EPI_ISL_402121
Original
2019-12-30
Human National Institute for Viral Disease Control and Prevention, China CDC
BetaCoV/Wuhan/IPBCAMS-WH-01/2019 EPI_ISL_402123
Original
2019-12-24
Human Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Pel
BetaCoV/Wuhan/WIV04/2019
EPLISL_402124 Original
2019-12-30
Human Wuhan Jinyintan Hospital](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:yqxcc5ayxllwkvel4narbha4/bafkreia5y3lwtbhh4ae7bctupomboyewluytybweememcbwpr6n42jxnfq@jpeg)

![screenshot of Eddie Holmes' tweet:
@edwardcholmes
All, an initial genome sequence of the coronavirus associated with the Wuhan outbreak is now available at Virological.org here: [link to virological]](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:yqxcc5ayxllwkvel4narbha4/bafkreif5tqxzjq4jcbdixilkvf74b626h34s2voszstutwib2wgtk5wkqa@jpeg)