www.nature.com/articles/s41...

www.nature.com/articles/s41...
What started as a shared vision for interoperable single-cell analysis has become a vibrant, global community.
From AnnData to full multimodal pipelines, we’re building the future of everything single-cell and spatial omics, together.
Here’s to what’s next!
What started as a shared vision for interoperable single-cell analysis has become a vibrant, global community.
From AnnData to full multimodal pipelines, we’re building the future of everything single-cell and spatial omics, together.
Here’s to what’s next!
Call for abstracts and registrations coming soon!

Call for abstracts and registrations coming soon!
Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...

Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...
Quite an indictment of some of the current single cell "virtual cell" foundation models. Even for the relatively mundane applications, cell labeling, batch correction etc, they are poor compared to much simpler & cheaper methods.
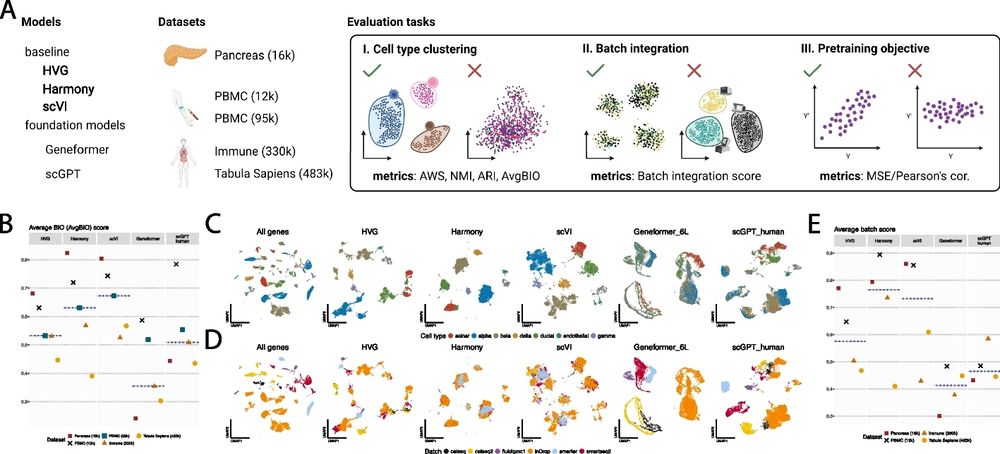
Quite an indictment of some of the current single cell "virtual cell" foundation models. Even for the relatively mundane applications, cell labeling, batch correction etc, they are poor compared to much simpler & cheaper methods.
July 18 or 19, 2025, Vancouver, Canada
Submit papers on OSS libraries, maintenance, best practices & more.
Format: 4-page non-archival papers
Due: May 19
codeml-workshop.github.io/codeml2025/#...

July 18 or 19, 2025, Vancouver, Canada
Submit papers on OSS libraries, maintenance, best practices & more.
Format: 4-page non-archival papers
Due: May 19
codeml-workshop.github.io/codeml2025/#...
Here’s (almost) everything we know about it
erictopol.substack.com/p/the-breakt...
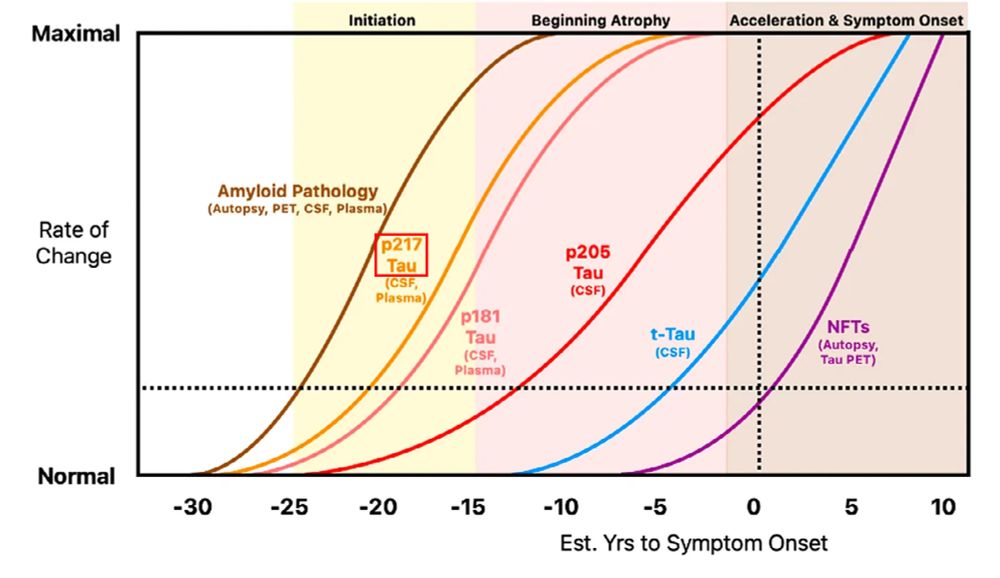
Here’s (almost) everything we know about it
erictopol.substack.com/p/the-breakt...

This is probably my most important paper. To my deep chagrin, it has no math.
XIST is a non-coding RNA exclusive to XX females. It silences one of the X chromosomes.
So what is it doing in male heart Schwann cells?
This is probably my most important paper. To my deep chagrin, it has no math.
XIST is a non-coding RNA exclusive to XX females. It silences one of the X chromosomes.
So what is it doing in male heart Schwann cells?
Thanks to @statnews.com naming me to STATUS List 2025 honoring leaders in health, medicine, and science!
#STATUSList
www.statnews.com/status-list/...

Thanks to @statnews.com naming me to STATUS List 2025 honoring leaders in health, medicine, and science!
#STATUSList
www.statnews.com/status-list/...
www.biorxiv.org/content/10.1...
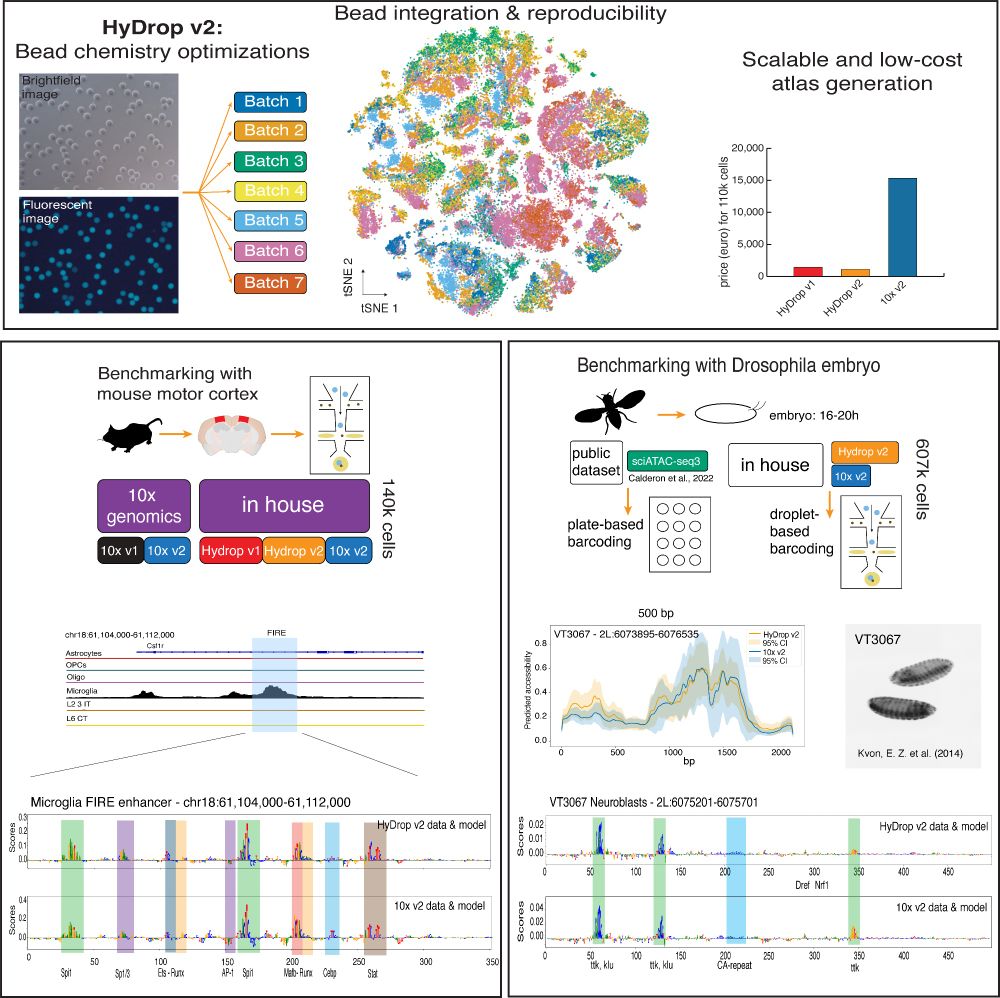
www.biorxiv.org/content/10.1...


Modelling and design of transcriptional enhancers
www.nature.com/articles/s44...

Modelling and design of transcriptional enhancers
www.nature.com/articles/s44...
Congrats: Anna, @xinmingtu.bsky.social , @lxsasse.bsky.social

Congrats: Anna, @xinmingtu.bsky.social , @lxsasse.bsky.social
Attend the premier conference at the intersection of ML & Bio, share your research and make lasting connections!
Submission deadline: June 1
More details: mlcb.github.io
Help spread the word—please RT! #MLCB2025
Attend the premier conference at the intersection of ML & Bio, share your research and make lasting connections!
Submission deadline: June 1
More details: mlcb.github.io
Help spread the word—please RT! #MLCB2025
Attend the premier conference at the intersection of ML & Bio, share your research and make lasting connections!
Submission deadline: June 1
More details: mlcb.github.io
Help spread the word—please RT! #MLCB2025
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


