
Department of Genetics, University of Cambridge. Wellcome Trust CDA fellow
https://www.gen.cam.ac.uk/research/research-groups/hocher-group
www.nature.com/articles/s41...




www.nature.com/articles/s41...
PhD /PostDoc positions available. Exploration of physiological functions of the cell fusogen Fusexin1 in Archaea.
In a collaborative project between the Albers lab (Freiburg) and the Podbilewicz lab (Technion) starting in January 2026.

PhD /PostDoc positions available. Exploration of physiological functions of the cell fusogen Fusexin1 in Archaea.
In a collaborative project between the Albers lab (Freiburg) and the Podbilewicz lab (Technion) starting in January 2026.
A collaborative effort spanning many years and several labs to uncover what the germline chromosomes of Paramecium really look like. 🔗 www.biorxiv.org/content/10.1...
1/5

A collaborative effort spanning many years and several labs to uncover what the germline chromosomes of Paramecium really look like. 🔗 www.biorxiv.org/content/10.1...
1/5
🌐 evedesign.bio
🌐 evedesign.bio
Today, we report the discovery of telomerase homologs in a family of antiviral RTs, revealing an unexpected evolutionary origin in bacteria.
www.biorxiv.org/content/10.1...

Today, we report the discovery of telomerase homologs in a family of antiviral RTs, revealing an unexpected evolutionary origin in bacteria.
www.biorxiv.org/content/10.1...
doi.org/10.1073/pnas...

doi.org/10.1073/pnas...

doi.org/10.1073/pnas...
www.nature.com/articles/s41...
Thanks again to our awesome collaborators @mblokesch.bsky.social and David and co and Mark Szczelkun and @steven-shaw.bsky.social and the DCI Lausanne @fbm-unil.bsky.social
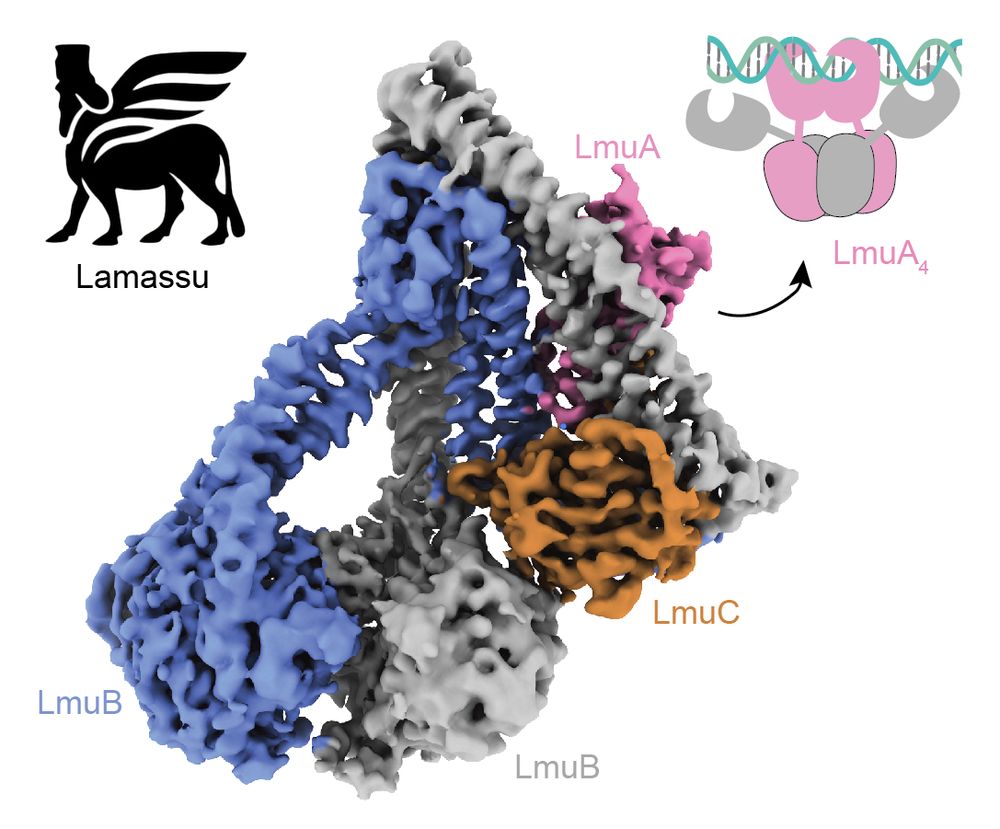
www.nature.com/articles/s41...
Thanks again to our awesome collaborators @mblokesch.bsky.social and David and co and Mark Szczelkun and @steven-shaw.bsky.social and the DCI Lausanne @fbm-unil.bsky.social

A fantastic collaboration with Antoine, with Jovana Kaljevic' initiated the collaboration and drives the project.

Repost = nice. Thank you very much!!!
Candidates who win Fellowships will be offered a Tenure Track Group Leader position from the outset, initially for 5 years.
Find out more: www.jic.ac.uk/training-car...

Repost = nice. Thank you very much!!!
Intergenic, repetitive motifs that act as nucleosome repleted regions in the budding yeast genome, have evolved de novo into novel protein-coding genes:
doi.org/10.1093/jeb/...
@vakirlis.bsky.social
@timothyfuqua.bsky.social

Intergenic, repetitive motifs that act as nucleosome repleted regions in the budding yeast genome, have evolved de novo into novel protein-coding genes:
doi.org/10.1093/jeb/...
@vakirlis.bsky.social
@timothyfuqua.bsky.social

🕰️We chart the full, synchronized lifecycle of a predatory bacterium using Hi-C, RNA-seq, ChIP-seq & microscopy.
🧬We uncover dramatic shifts in nucleoid architecture & transcription, tightly coordinated with predation.
www.biorxiv.org/content/10.1...

🕰️We chart the full, synchronized lifecycle of a predatory bacterium using Hi-C, RNA-seq, ChIP-seq & microscopy.
🧬We uncover dramatic shifts in nucleoid architecture & transcription, tightly coordinated with predation.
www.biorxiv.org/content/10.1...

@rkoszul.bsky.social @natmicrobiol.nature.com @cnrs.fr @institutpasteur.bsky.social
www.nature.com/articles/s41...

@rkoszul.bsky.social @natmicrobiol.nature.com @cnrs.fr @institutpasteur.bsky.social
www.nature.com/articles/s41...



