Arne Schneuing
@rne.bsky.social
680 followers
140 following
9 posts
PhD student @EPFL 🇨🇭
ML & computational biology 🤖🧬⚛️
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Arne Schneuing
Reposted by Arne Schneuing
Nature
@nature.com
· Sep 1

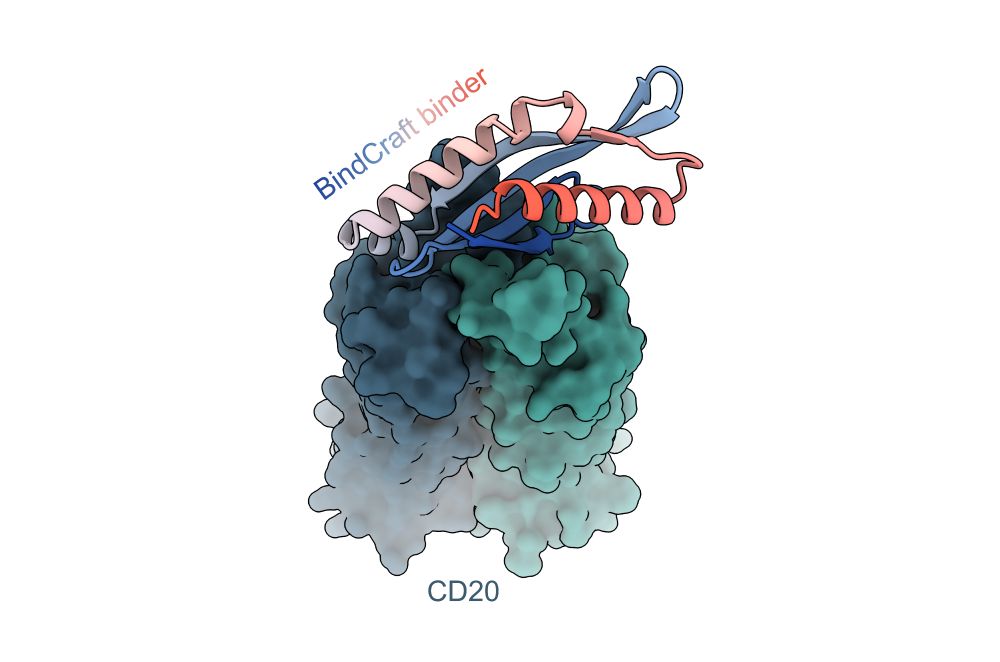
One-shot design of functional protein binders with BindCraft - Nature
BindCraft, an open-source, automated pipeline for de novo protein binder design with experimental success rates of 10–100%, leverages AlphaFold2 weights to generate binders with nanomolar affinity without the need for high-throughput screening.
go.nature.com
Reposted by Arne Schneuing
Reposted by Arne Schneuing
Reposted by Arne Schneuing
Frank Noe
@franknoe.bsky.social
· Jul 10
Reposted by Arne Schneuing
Reposted by Arne Schneuing
Reposted by Arne Schneuing
Reposted by Arne Schneuing
Rebecca Neeser
@rebeccaneeser.bsky.social
· Apr 24

Flow-Based Fragment Identification via Contrastive Learning of...
Fragment-based drug design is a promising strategy leveraging the binding of individual fragments, potentially yielding ligands with multiple key interactions, surpassing the efficiency of full...
openreview.net
Reposted by Arne Schneuing
Reposted by Arne Schneuing
Reposted by Arne Schneuing
Reposted by Arne Schneuing
Reposted by Arne Schneuing
Arne Schneuing
@rne.bsky.social
· Mar 7
Reposted by Arne Schneuing
Frank Noe
@franknoe.bsky.social
· Feb 19
Reposted by Arne Schneuing
Arne Schneuing
@rne.bsky.social
· Jan 15

Targeting protein-ligand neosurfaces using a generalizable deep learning approach
Molecular recognition events between proteins drive biological processes in living systems. However, higher levels of mechanistic regulation have emerged, where protein-protein interactions are condit...
biorxiv.org