Boyi Guo (郭博逸)
@boyiguo.bsky.social
150 followers
120 following
14 posts
Biomedical data scientist working at the intersection of machine learning 🤖, computational omics 🧬, and population health 👨👩👧👦
👨💻 Incoming assistant professor @University of Utah
🔗 Personal website: https://boyi-guo.com/
👤 Pronounce: he/him
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Boyi Guo (郭博逸)
Rita Strack
@ritastrack.bsky.social
· Jul 9
Reposted by Boyi Guo (郭博逸)
Rita Strack
@ritastrack.bsky.social
· Jul 9
Boyi Guo (郭博逸)
@boyiguo.bsky.social
· Jun 24
Boyi Guo (郭博逸)
@boyiguo.bsky.social
· Jun 24

Genomics of schizophrenia, bipolar disorder and major depressive disorder
Nature Reviews Genetics - Genomic advances have enhanced our understanding of schizophrenia, bipolar disorder and major depressive disorder, revealing genetic architectures and risk mechanisms...
www.nature.com
Reposted by Boyi Guo (郭博逸)
Reposted by Boyi Guo (郭博逸)
Boyi Guo (郭博逸)
@boyiguo.bsky.social
· Jun 10
Boyi Guo (郭博逸)
@boyiguo.bsky.social
· Jun 10
Reposted by Boyi Guo (郭博逸)
Michael Totty
@mictott.bsky.social
· Jun 6
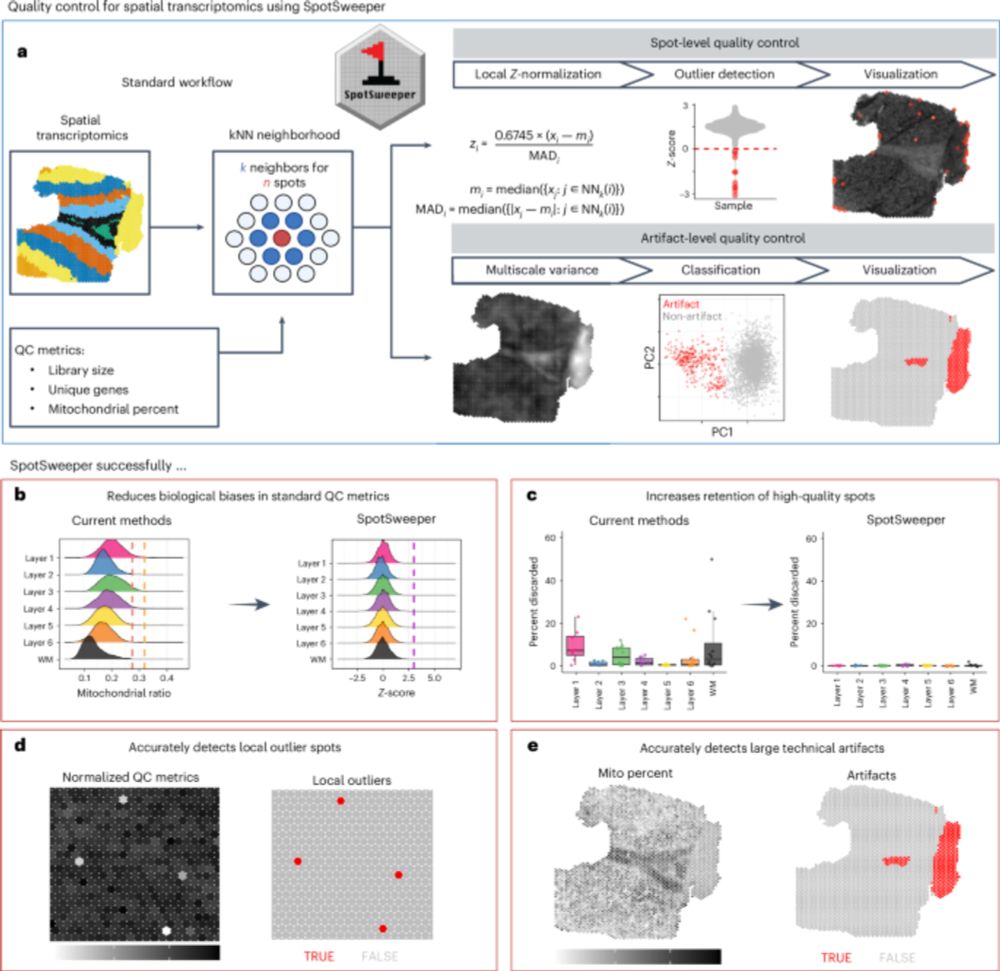
SpotSweeper: spatially aware quality control for spatial transcriptomics - Nature Methods
SpotSweeper is a spatially aware method for quality control of spatially resolved transcriptomics data that corrects for spatial confounding missed by existing methods, including both local and region...
nature.com
Boyi Guo (郭博逸)
@boyiguo.bsky.social
· Jun 6
Nature Methods
@natmethods.nature.com
· Jun 6

SpotSweeper: spatially aware quality control for spatial transcriptomics - Nature Methods
SpotSweeper is a spatially aware method for quality control of spatially resolved transcriptomics data that corrects for spatial confounding missed by existing methods, including both local and region...
www.nature.com
Reposted by Boyi Guo (郭博逸)
Rita Strack
@ritastrack.bsky.social
· Apr 10
Boyi Guo (郭博逸)
@boyiguo.bsky.social
· Dec 5
Reposted by Boyi Guo (郭博逸)
Reposted by Boyi Guo (郭博逸)
Kinnary Shah
@kinnaryshah.bsky.social
· Nov 16
Boyi Guo (郭博逸)
@boyiguo.bsky.social
· Dec 29
Boyi Guo (郭博逸)
@boyiguo.bsky.social
· Oct 23
Boyi Guo (郭博逸)
@boyiguo.bsky.social
· Oct 23
Boyi Guo (郭博逸)
@boyiguo.bsky.social
· Oct 20
Boyi Guo (郭博逸)
@boyiguo.bsky.social
· Oct 2