
BS-MS, Mahipal Ganji lab, IISc
SMCs | NAPs | genome organization | DNA repair 🧬
www.biorxiv.org/content/10.1...

We show how cohesin integrates loop extrusion with sister tethering to guide the homology search during DNA repair.
Huge congratulations to all authors, and to @fedeteloni.bsky.social for leading this work 👏
We’re excited to see his next steps!
Our study reveals how cohesin guides focused and accurate homology search.
Read more 👉 www.science.org/doi/10.1126/...
Follow along for key insights and updates! 🧵

We show how cohesin integrates loop extrusion with sister tethering to guide the homology search during DNA repair.
Huge congratulations to all authors, and to @fedeteloni.bsky.social for leading this work 👏
We’re excited to see his next steps!
👉 www.molgen.mpg.de/5098010/stei...

👉 www.molgen.mpg.de/5098010/stei...
This prestigious grant allows Leonid Mirny (MIT), Wendy Bickmore (Edinburgh), Benjamin Rowland (NKI) and me to study the DNA-looping SMC motors proteins that control our chromosomes in the next 6 years
Sixty-six research teams have been selected for funding, bringing together 239 scientists. Congratulations to all!
➡️ buff.ly/PSn3bi9
#EUfunded #HorizonEurope #ERCSyG
This prestigious grant allows Leonid Mirny (MIT), Wendy Bickmore (Edinburgh), Benjamin Rowland (NKI) and me to study the DNA-looping SMC motors proteins that control our chromosomes in the next 6 years
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...

Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...
www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇
academic.oup.com/nar/article/... @narjournal.bsky.social

academic.oup.com/nar/article/... @narjournal.bsky.social
We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...

We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...
I currently have 5 (!) open positions for postdocs/PhD students in our CDlab for projects:
- Nanopore protein sequencing
- Archaeal CDV cell division
- Microfluidics for synthetic cells
- Nuclear Pore Complex
- Origami mimics of peroxisomes
Please apply! ceesdekkerlab.nl/come-join-us/
RT=👍

I currently have 5 (!) open positions for postdocs/PhD students in our CDlab for projects:
- Nanopore protein sequencing
- Archaeal CDV cell division
- Microfluidics for synthetic cells
- Nuclear Pore Complex
- Origami mimics of peroxisomes
Please apply! ceesdekkerlab.nl/come-join-us/
RT=👍
Thrilled to share that we have discovered a brand-new anti-phage defense system! Bacteria have evolved various defense strategies (CRISPR etc) to counter phage attacks. We found a new one - fascinating and dramatic
⚔️🦠❄️🔬
Thrilled to share that we have discovered a brand-new anti-phage defense system! Bacteria have evolved various defense strategies (CRISPR etc) to counter phage attacks. We found a new one - fascinating and dramatic
⚔️🦠❄️🔬
🧬 Fully funded PhD opportunity! 🧬
We are looking for a curious and motivated candidate to join our new team in Utrecht. Our goal is to develop biomimetic mechanisms for protein folding using DNA origami.
More info:
🔗 www.uu.nl/en/organisat...
🗓️ Deadline: 15.08
🧬 Fully funded PhD opportunity! 🧬
We are looking for a curious and motivated candidate to join our new team in Utrecht. Our goal is to develop biomimetic mechanisms for protein folding using DNA origami.
More info:
🔗 www.uu.nl/en/organisat...
🗓️ Deadline: 15.08
#nanopores #smFRET #biomolecular #dynamics #singleMolecules
We’re looking for talented & ambitious new colleagues enthusiastic about biomolecular dynamics & single-molecule tech.
Please share broadly, thank you!🤝
schmid.chemie.unibas.ch
#nanopores #smFRET #biomolecular #dynamics #singleMolecules
We’re looking for talented & ambitious new colleagues enthusiastic about biomolecular dynamics & single-molecule tech.
Please share broadly, thank you!🤝
schmid.chemie.unibas.ch
If you find it interesting, please repost it!
#science #scicomm #education




If you find it interesting, please repost it!
#science #scicomm #education
www.biorxiv.org/content/10.1...
1/
www.biorxiv.org/content/10.1...
1/
In an exemplary in vitro/in vivo study, 1st authors @btanalikwu.bsky.social & Alice Deshayes showed that dense linear protein arrays (e.g. telomeres) do stop loop-extruding SMCs!
Great collaboration with Marcand lab.
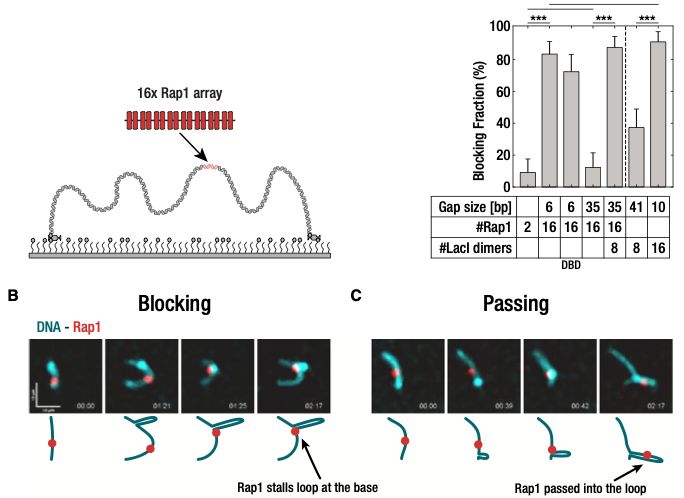
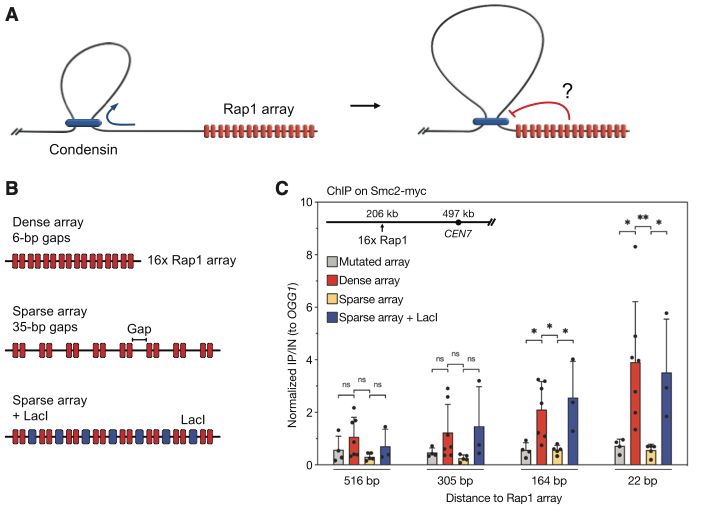
In an exemplary in vitro/in vivo study, 1st authors @btanalikwu.bsky.social & Alice Deshayes showed that dense linear protein arrays (e.g. telomeres) do stop loop-extruding SMCs!
Great collaboration with Marcand lab.
This short article sketches the state of the art of the field that studies the structure and dynamic gating mechanism of the nuclear pore complex


This short article sketches the state of the art of the field that studies the structure and dynamic gating mechanism of the nuclear pore complex
Q: how does chromatin move?
Using MINFLUX, SPT & SRLCI, we track chromatin dynamics across 7 orders of magnitude in time to provide answers www.biorxiv.org/content/10.1...

Q: how does chromatin move?
Using MINFLUX, SPT & SRLCI, we track chromatin dynamics across 7 orders of magnitude in time to provide answers www.biorxiv.org/content/10.1...
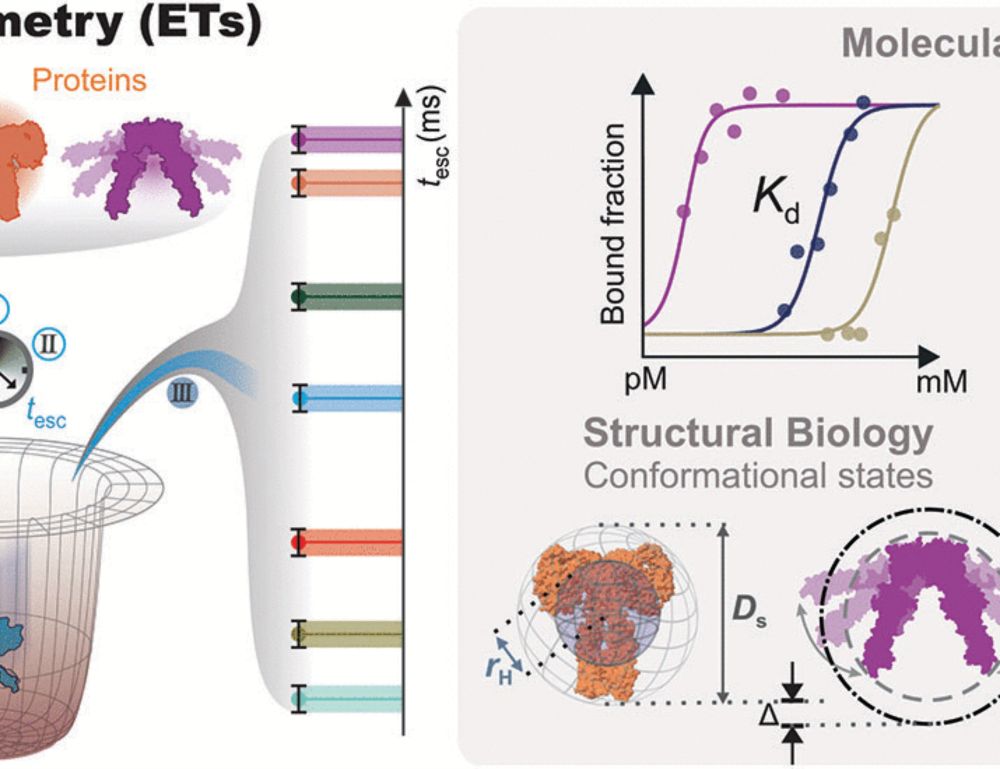
We can directly quantify stoichiometry and oligomerization from super-res (DNA-PAINT, RESI) images!! 🧬🎨
We are excited to present our latest work published in @natcomms.nature.com
www.nature.com/articles/s41...
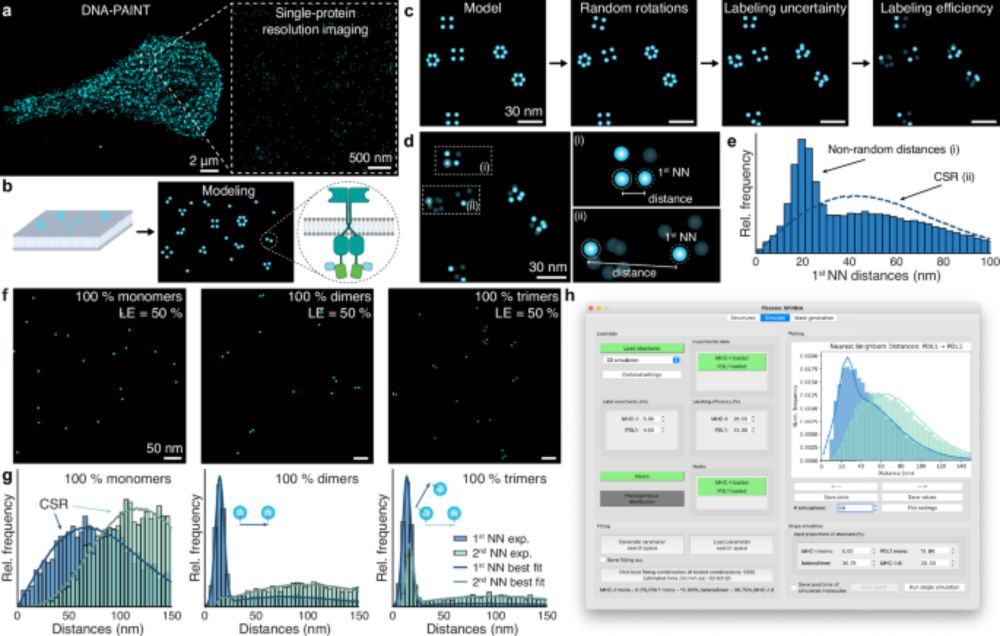
We can directly quantify stoichiometry and oligomerization from super-res (DNA-PAINT, RESI) images!! 🧬🎨
Fan Fang Varshini Ramanathan in collab w Jie Liu.
Q: How do we get ultra-high-res 3D genome maps?
A: New deep learning model, Cleopatra.
Cleo trains on Micro-C, fine-tunes on RCMC, and predicts genome-wide 3D maps at ultra-high resolution.




Fan Fang Varshini Ramanathan in collab w Jie Liu.
Q: How do we get ultra-high-res 3D genome maps?
A: New deep learning model, Cleopatra.
Cleo trains on Micro-C, fine-tunes on RCMC, and predicts genome-wide 3D maps at ultra-high resolution.
Lipid flip flop regulates the shape of growing and dividing synthetic cells
www.biorxiv.org/content/10.1...
Here, postdoc Rafa Lira studied GUV growth by SUV fusions to show that lipid flip flop is key for low curvature stress and symmetric buds


Lipid flip flop regulates the shape of growing and dividing synthetic cells
www.biorxiv.org/content/10.1...
Here, postdoc Rafa Lira studied GUV growth by SUV fusions to show that lipid flip flop is key for low curvature stress and symmetric buds
www.science.org/doi/10.1126/...
A short clip describing the key results:
www.youtube.com/watch?v=pmvO...

www.science.org/doi/10.1126/...
A short clip describing the key results:
www.youtube.com/watch?v=pmvO...
www.biochemistry.org/about-us/new...
It is wonderful that the Biochemical Society honors our contributions to single-molecule biophysics in this way!

www.biochemistry.org/about-us/new...
It is wonderful that the Biochemical Society honors our contributions to single-molecule biophysics in this way!
pdb101.rcsb.org/sci-art/bezs...
@rcsbpdb.bsky.social
@pdbeurope.bsky.social
#sciart
pdb101.rcsb.org/sci-art/bezs...
@rcsbpdb.bsky.social
@pdbeurope.bsky.social
#sciart
Here, 1st authors Alex Joesaar & Martin Holub developed a microfluidics platform to extract and study full length megabase-size genomes in microfluidic chambers (where they are protected against shearing by flow)!




Here, 1st authors Alex Joesaar & Martin Holub developed a microfluidics platform to extract and study full length megabase-size genomes in microfluidic chambers (where they are protected against shearing by flow)!

