
PhD candidate with the Pritchard Lab @Stanford

www.biorxiv.org/content/10.1...
(1/n)

www.biorxiv.org/content/10.1...
(1/n)
I am looking for a computational scientist to join my genomics lab at Stanford. They should have an outstanding skillset in ML/statistical methods for genomic applications, postdoc experience and a strong publication record.
#sciencejobs
I am looking for a computational scientist to join my genomics lab at Stanford. They should have an outstanding skillset in ML/statistical methods for genomic applications, postdoc experience and a strong publication record.
#sciencejobs

I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵
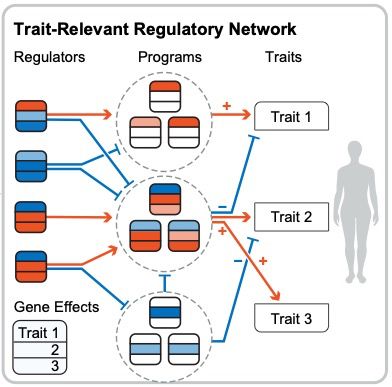
I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵

Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...

Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...
For many traits the number of duplications or loss-of-function (LoF) mutations is correlated with phenotype. Curiously, for most traits, the AVERAGE direction of LoFs and Dups is the SAME. Why?
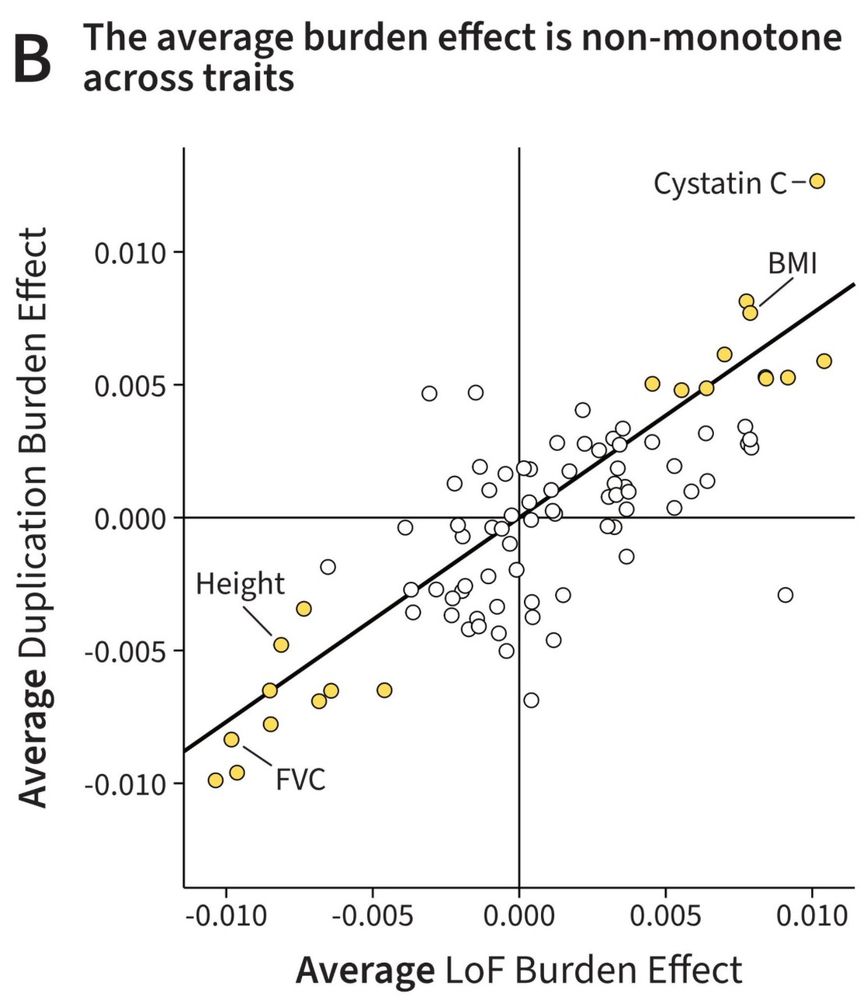
For many traits the number of duplications or loss-of-function (LoF) mutations is correlated with phenotype. Curiously, for most traits, the AVERAGE direction of LoFs and Dups is the SAME. Why?
Here's a link to my notes on population & quantitative genetics:
github.com/cooplab/popg...
Hoping to extend it more after the winter holidays, as I'm just finishing up teaching the undergrad version of class.
Here's a link to my notes on population & quantitative genetics:
github.com/cooplab/popg...
Hoping to extend it more after the winter holidays, as I'm just finishing up teaching the undergrad version of class.
🧵(1/8)

🧵(1/8)

