Interested in complex trait genetics and immunology.
We show how data on expression quantitative trait loci (eQTL) relates to the structure of gene regulatory networks (GRN). Much of the GRN / eQTL picture is unmapped, but what we do have says a lot… (1/)
doi.org/10.1101/2025...
We show how data on expression quantitative trait loci (eQTL) relates to the structure of gene regulatory networks (GRN). Much of the GRN / eQTL picture is unmapped, but what we do have says a lot… (1/)
doi.org/10.1101/2025...
We'll work at the intersection of statistical genetics, population genetics, and machine learning.
We'll work at the intersection of statistical genetics, population genetics, and machine learning.
Email CV to me+cc my assistant (see 'contact' on my website). Ad to follow.
Email CV to me+cc my assistant (see 'contact' on my website). Ad to follow.
While this seems at odds with our recent work about how burden tests and GWAS prioritize different genes, our results agree (🧬🧪🧵 1/6)
www.biorxiv.org/content/10.1...

While this seems at odds with our recent work about how burden tests and GWAS prioritize different genes, our results agree (🧬🧪🧵 1/6)
www.biorxiv.org/content/10.1...
www.cell.com/cell-genomic...

www.cell.com/cell-genomic...
www.science.org/doi/10.1126/...
TL;DR: BCRs ARE ALL YOU NEED!
(Well actually .... keep reading) 1/

www.science.org/doi/10.1126/...
TL;DR: BCRs ARE ALL YOU NEED!
(Well actually .... keep reading) 1/
Research leaders call for an end to substantial underfunding of interdisciplinary research in Japan.
On my current visit to 🇯🇵 I can see the country is ready for a change
#japan #academicSky 🧪
www.nature.com/articles/d41...

Research leaders call for an end to substantial underfunding of interdisciplinary research in Japan.
On my current visit to 🇯🇵 I can see the country is ready for a change
#japan #academicSky 🧪
www.nature.com/articles/d41...
Here I want to expand on the value of using DIRECTIONAL information contained in LoF burden tests.🧵
[work led by @minetoota.bsky.social ]
bsky.app/profile/jkpr...
But interpretation is difficult as most effects flow through (unobserved) gene regulatory networks. Can we gain insight by linking to modern Perturb-seq data?
www.biorxiv.org/content/10.1...

Here I want to expand on the value of using DIRECTIONAL information contained in LoF burden tests.🧵
[work led by @minetoota.bsky.social ]
bsky.app/profile/jkpr...
I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵
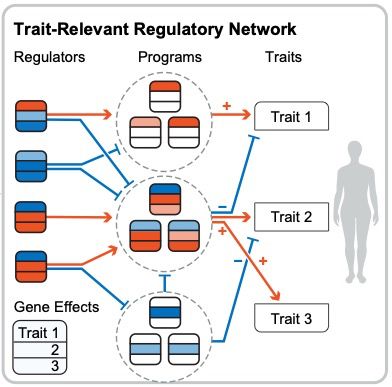
combining ‘quantitative estimates of gene-trait relationships from loss-of-function burden tests with gene-regulatory connections inferred from Perturb-seq experiments in relevant cell types’ 👇
I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵

combining ‘quantitative estimates of gene-trait relationships from loss-of-function burden tests with gene-regulatory connections inferred from Perturb-seq experiments in relevant cell types’ 👇
I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵

Allows me to reuse one of my figures from a few weeks ago on a gene as old as eukaryotes, mitoferrin, which is needed to move iron into mitochondria.
@jkpritch.bsky.social
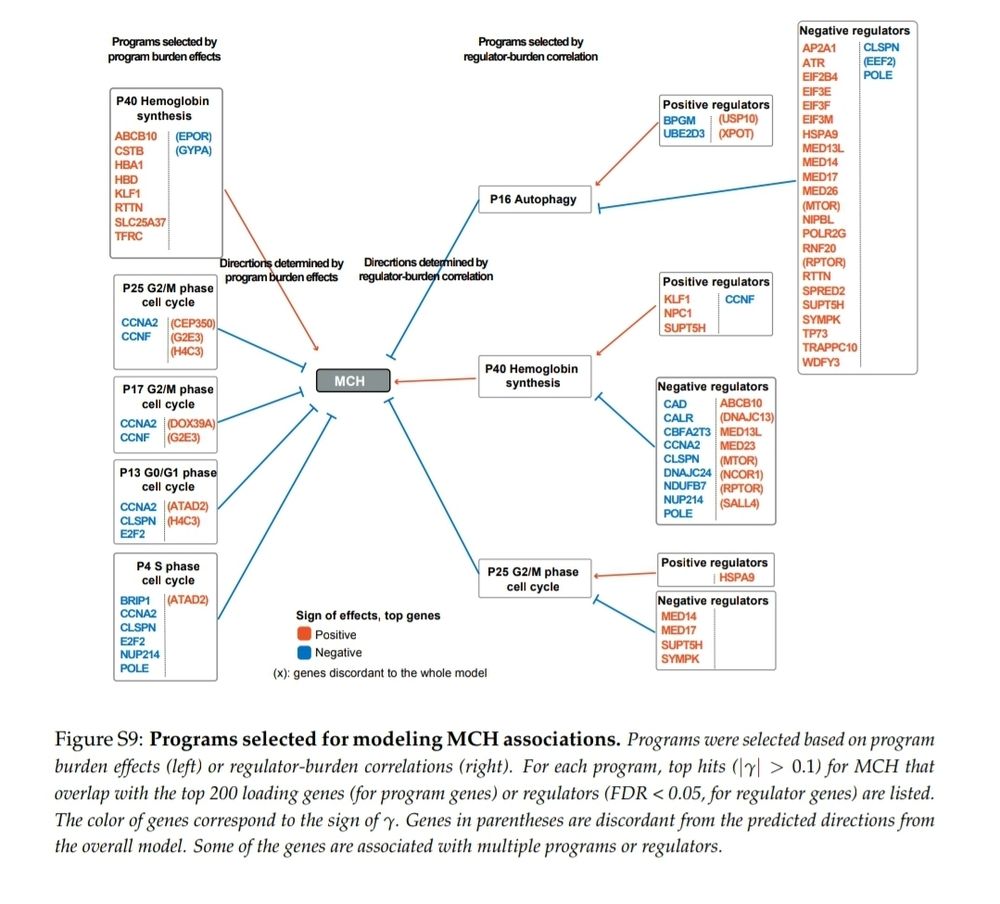

Allows me to reuse one of my figures from a few weeks ago on a gene as old as eukaryotes, mitoferrin, which is needed to move iron into mitochondria.
@jkpritch.bsky.social
We dove into how we can model the gene regulatory architecture of complex traits with 1) Gene effects from LoF burden tests and 2) Perturb-seq.
I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵

We dove into how we can model the gene regulatory architecture of complex traits with 1) Gene effects from LoF burden tests and 2) Perturb-seq.
A quick thread about the importance of thinking about all traits at once 👇 1/6 (🧪🧬)
A quick thread about the importance of thinking about all traits at once 👇 1/6 (🧪🧬)
Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...

Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...
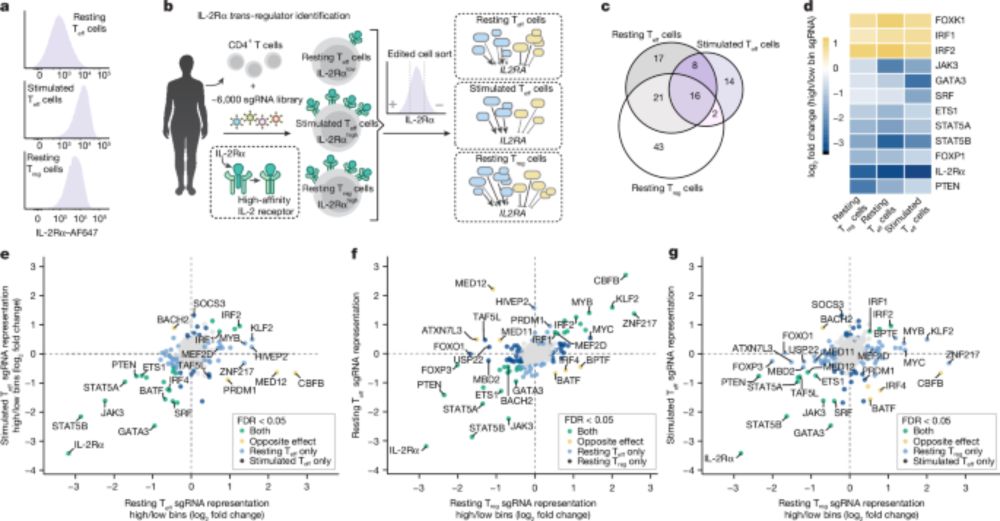
www.nature.com/articles/s41...

