📍 @ml4science.bsky.social, Tübingen, Germany
IV-9: @stewah.bsky.social presents joint work with @danielged.bsky.social: A new perspective on LLM-based model discovery with applications in neuroscience 6/9
IV-9: @stewah.bsky.social presents joint work with @danielged.bsky.social: A new perspective on LLM-based model discovery with applications in neuroscience 6/9
III-6. @matthijspals.bsky.social will present: Sequence memory in distinct subspaces in data-constrained RNNs of human working memory (joint work with @stefanieliebe.bsky.social) 4/9
III-6. @matthijspals.bsky.social will present: Sequence memory in distinct subspaces in data-constrained RNNs of human working memory (joint work with @stefanieliebe.bsky.social) 4/9
II-4. Isaac Omolayo will present: Contrastive Learning for Predicting Neural Activity in Connectome Constrained Deep Mechanistic Networks 2/9
II-4. Isaac Omolayo will present: Contrastive Learning for Predicting Neural Activity in Connectome Constrained Deep Mechanistic Networks 2/9
@ml4science.bsky.social, @tuebingen-ai.bsky.social, @unituebingen.bsky.social
@ml4science.bsky.social, @tuebingen-ai.bsky.social, @unituebingen.bsky.social
💻 Code is available: github.com/mackelab/npe...

💻 Code is available: github.com/mackelab/npe...
By leveraging foundation models like TabPFN, we can make SBI training-free, simulation-efficient, and easy to use.
This work is another step toward user-friendly Bayesian inference for a broader science and engineering community.
By leveraging foundation models like TabPFN, we can make SBI training-free, simulation-efficient, and easy to use.
This work is another step toward user-friendly Bayesian inference for a broader science and engineering community.
🧠 single-compartment neuron
🦀 31-parameter crab pyloric network
NPE-PF delivers tight posteriors & accurate predictions with far fewer simulations than previous methods.
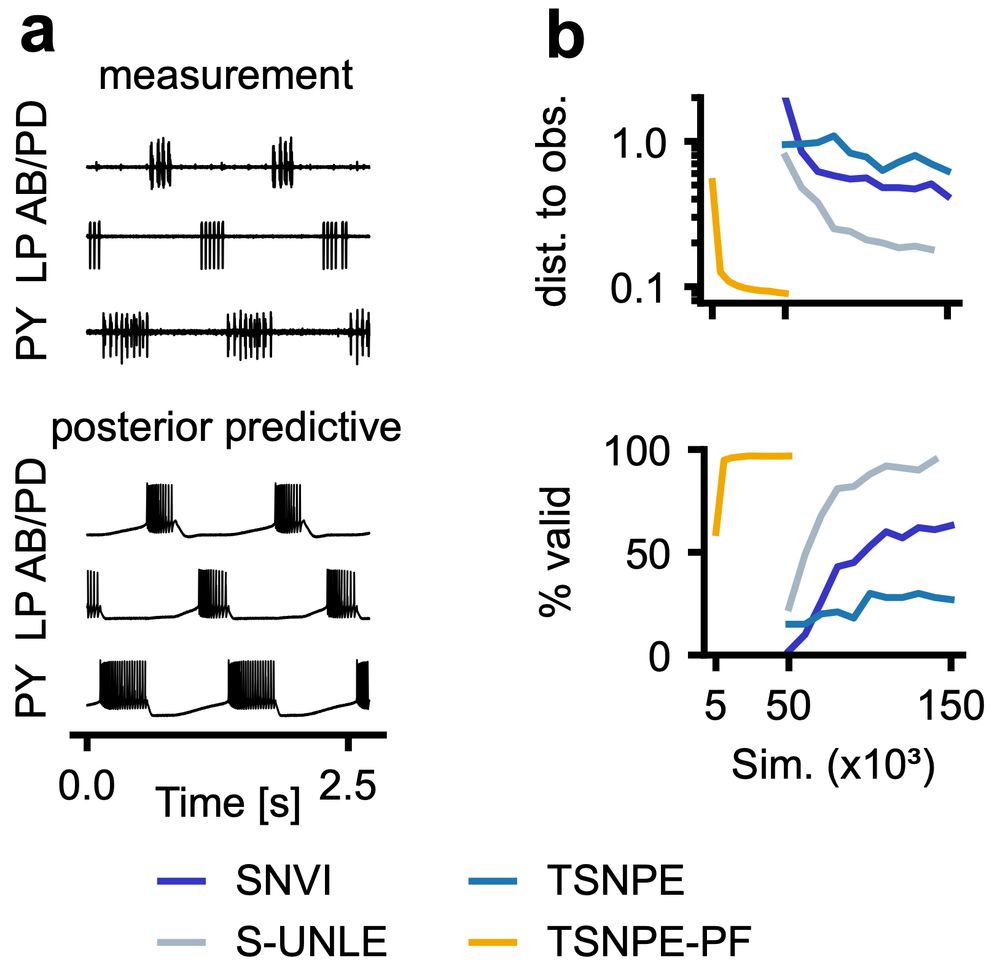
🧠 single-compartment neuron
🦀 31-parameter crab pyloric network
NPE-PF delivers tight posteriors & accurate predictions with far fewer simulations than previous methods.
🚫 No need to train inference nets or tune hyperparameters.
🌟 Competitive or superior performance vs. standard SBI methods.
🚀 Especially strong performance for smaller simulation budgets.
🔄 Filtering to handle large datasets + support for sequential inference.
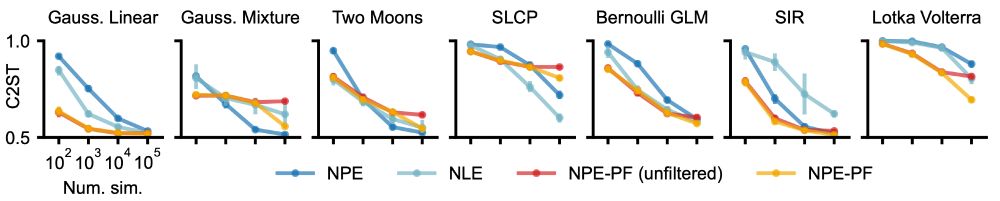
🚫 No need to train inference nets or tune hyperparameters.
🌟 Competitive or superior performance vs. standard SBI methods.
🚀 Especially strong performance for smaller simulation budgets.
🔄 Filtering to handle large datasets + support for sequential inference.
TabPFN, originally trained for tabular regression and classification, can estimate posteriors by autoregressively modeling one parameter dimension after the other.
It’s remarkably effective, even though TabPFN was not designed for SBI.
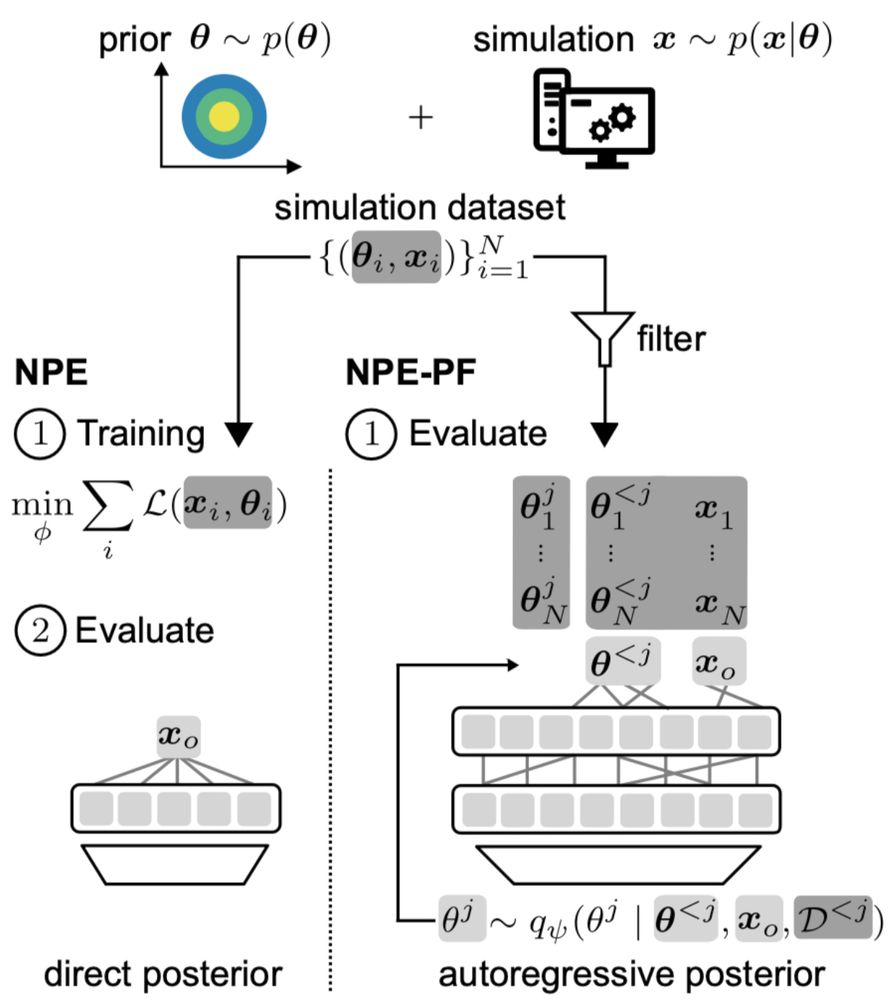
TabPFN, originally trained for tabular regression and classification, can estimate posteriors by autoregressively modeling one parameter dimension after the other.
It’s remarkably effective, even though TabPFN was not designed for SBI.
⚠️ Simulators can be expensive
⚠️ Training & tuning neural nets can be tedious
Our method NPE-PF repurposes TabPFN as an in-context density estimator for training-free, simulation-efficient Bayesian inference.
⚠️ Simulators can be expensive
⚠️ Training & tuning neural nets can be tedious
Our method NPE-PF repurposes TabPFN as an in-context density estimator for training-free, simulation-efficient Bayesian inference.
Code: github.com/mackelab/sbi...
Paper: www.cambridge.org/core/journal...
Code: github.com/mackelab/sbi...
Paper: www.cambridge.org/core/journal...