
Immune proteins.
Huge thanks to everyone who made it happen 💙 - especially my brilliant collaborators
David W. Adams, Eliane Hadas Yardeni, @mblokesch.bsky.social, and of course @soreklab.bsky.social
Huge thanks to everyone who made it happen 💙 - especially my brilliant collaborators
David W. Adams, Eliane Hadas Yardeni, @mblokesch.bsky.social, and of course @soreklab.bsky.social
This strategy could:
✅ Broaden the host range of therapeutic phages.
✅ Allow personalized phages for each patient’s infection.
✅ Unlock genetic engineering in “untouchable” bacteria for biotech + medicine.
This strategy could:
✅ Broaden the host range of therapeutic phages.
✅ Allow personalized phages for each patient’s infection.
✅ Unlock genetic engineering in “untouchable” bacteria for biotech + medicine.
We designed inhibitors for DdmDE, a system in Vibrio cholerae that usually clears plasmids from transformed cells.
Two of these binders conferred the plasmids with significant resistance against DdmDE-mediated plasmid clearance even in its native bacterial strain.

We designed inhibitors for DdmDE, a system in Vibrio cholerae that usually clears plasmids from transformed cells.
Two of these binders conferred the plasmids with significant resistance against DdmDE-mediated plasmid clearance even in its native bacterial strain.
➡️ This phage could overcome two defenses in the same bacterium.
This shows we can build “multi-resistant” phages for therapy.

➡️ This phage could overcome two defenses in the same bacterium.
This shows we can build “multi-resistant” phages for therapy.
Synthetic binders blocked Avs1 activity—again, letting phages replicate in bacteria that co-express our binders with Avs1.

Synthetic binders blocked Avs1 activity—again, letting phages replicate in bacteria that co-express our binders with Avs1.


We hypothesized that designing small proteins that bind the molecule-sensing site in the SLOG domain would disrupt its ability to perceive the signaling molecules.
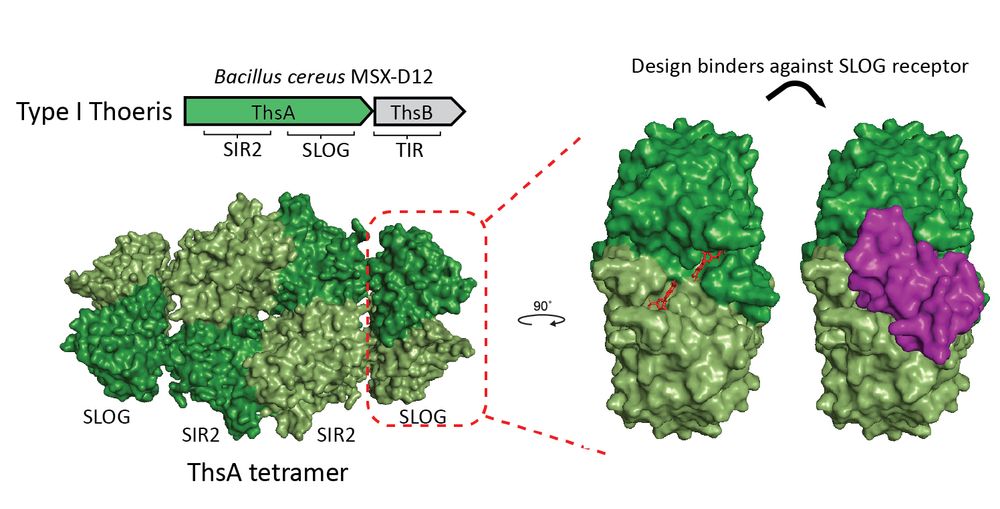
We hypothesized that designing small proteins that bind the molecule-sensing site in the SLOG domain would disrupt its ability to perceive the signaling molecules.
Using AI-driven de novo protein design (RFdiffusion), we created small proteins (49–98 aa) that bind and block bacterial defense proteins 💻🤖🧬
Using AI-driven de novo protein design (RFdiffusion), we created small proteins (49–98 aa) that bind and block bacterial defense proteins 💻🤖🧬
Bacteria are far from defenseless.
They carry dozens of anti-phage and anti-plasmid systems—CRISPR is just the tip of the iceberg.
➡️ This makes phage therapy often fail in patients.
➡️ It also makes many bacteria very hard to genetically engineer.
Bacteria are far from defenseless.
They carry dozens of anti-phage and anti-plasmid systems—CRISPR is just the tip of the iceberg.
➡️ This makes phage therapy often fail in patients.
➡️ It also makes many bacteria very hard to genetically engineer.

