
Immune proteins.
We designed inhibitors for DdmDE, a system in Vibrio cholerae that usually clears plasmids from transformed cells.
Two of these binders conferred the plasmids with significant resistance against DdmDE-mediated plasmid clearance even in its native bacterial strain.

We designed inhibitors for DdmDE, a system in Vibrio cholerae that usually clears plasmids from transformed cells.
Two of these binders conferred the plasmids with significant resistance against DdmDE-mediated plasmid clearance even in its native bacterial strain.
➡️ This phage could overcome two defenses in the same bacterium.
This shows we can build “multi-resistant” phages for therapy.

➡️ This phage could overcome two defenses in the same bacterium.
This shows we can build “multi-resistant” phages for therapy.
Synthetic binders blocked Avs1 activity—again, letting phages replicate in bacteria that co-express our binders with Avs1.

Synthetic binders blocked Avs1 activity—again, letting phages replicate in bacteria that co-express our binders with Avs1.


We hypothesized that designing small proteins that bind the molecule-sensing site in the SLOG domain would disrupt its ability to perceive the signaling molecules.
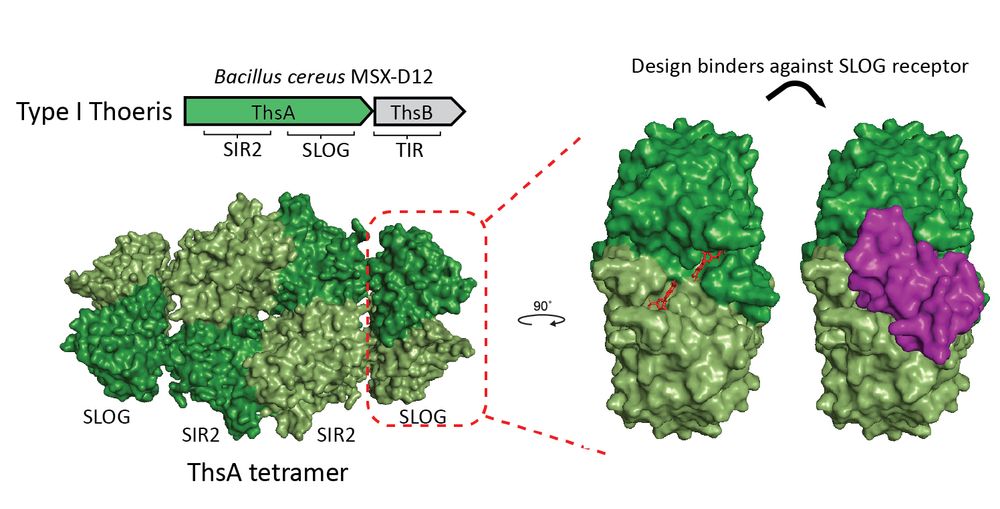
We hypothesized that designing small proteins that bind the molecule-sensing site in the SLOG domain would disrupt its ability to perceive the signaling molecules.

